Journal of Machine Learning Research 19 (2018) 1-30 Submitted 9/16; Revised 7/18; Published 9/18
Random Forests, Decision Trees, and Categorical Predictors:
The “Absent Levels” Problem
Google LLC
1600 Amphitheatre Parkway
Mountain View, CA 94043, USA
Editor: Sebastian Nowozin
Abstract
One advantage of decision tree based methods like random forests is their ability to natively
handle categorical predictors without having to first transform them (e.g., by using feature
engineering techniques). However, in this paper, we show how this capability can lead to
an inherent “absent levels” problem for decision tree based methods that has never been
thoroughly discussed, and whose consequences have never been carefully explored. This
problem occurs whenever there is an indeterminacy over how to handle an observation that
has reached a categorical split which was determined when the observation in question’s
level was absent during training. Although these incidents may appear to be innocuous, by
using Leo Breiman and Adele Cutler’s random forests FORTRAN code and the randomForest
R package (Liaw and Wiener, 2002) as motivating case studies, we examine how overlooking
the absent levels problem can systematically bias a model. Furthermore, by using three real
data examples, we illustrate how absent levels can dramatically alter a model’s performance
in practice, and we empirically demonstrate how some simple heuristics can be used to help
mitigate the effects of the absent levels problem until a more robust theoretical solution is
found.
Keywords: absent levels, categorical predictors, decision trees, CART, random forests
1. Introduction
Since its introduction in Breiman (2001), random forests have enjoyed much success as one
of the most widely used decision tree based methods in machine learning. But despite
their popularity and apparent simplicity, random forests have proven to be very difficult
to analyze. Indeed, many of the basic mathematical properties of the algorithm are still
not completely well understood, and theoretical investigations have often had to rely on
either making simplifying assumptions or considering variations of the standard framework
in order to make the analysis more tractable—see, for example, Biau et al. (2008), Biau
(2012), and Denil et al. (2014).
One advantage of decision tree based methods like random forests is their ability to
natively handle categorical predictors without having to first transform them (e.g., by using
feature engineering techniques). However, in this paper, we show how this capability can
lead to an inherent “absent levels” problem for decision tree based methods that has, to the
best of our knowledge, never been thoroughly discussed, and whose consequences have never
been carefully explored. This problem occurs whenever there is an indeterminacy over how
c
2018 Timothy C. Au.
License: CC-BY 4.0, see https://creativecommons.org/licenses/by/4.0/. Attribution requirements are provided
at http://jmlr.org/papers/v19/16-474.html.

Au
to handle an observation that has reached a categorical split which was determined when
the observation in question’s level was absent during training—an issue that can arise in
three different ways:
1. The levels are present in the population but, due to sampling variability, are absent
in the training set.
2. The levels are present in the training set but, due to bagging, are absent in an indi-
vidual tree’s bootstrapped sample of the training set.
3. The levels are present in an individual tree’s training set but, due to a series of earlier
node splits, are absent in certain branches of the tree.
These occurrences subsequently result in situations where observations with absent levels
are unsure of how to proceed further down the tree—an intrinsic problem for decision tree
based methods that has seemingly been overlooked in both the theoretical literature and in
much of the software that implements these methods.
Although these incidents may appear to be innocuous, by using Leo Breiman and Adele
Cutler’s random forests FORTRAN code and the randomForest R package (Liaw and Wiener,
2002) as motivating case studies,
1
we examine how overlooking the absent levels problem
can systematically bias a model. In addition, by using three real data examples, we il-
lustrate how absent levels can dramatically alter a model’s performance in practice, and
we empirically demonstrate how some simple heuristics can be used to help mitigate their
effects.
The rest of this paper is organized as follows. In Section 2, we introduce some notation
and provide an overview of the random forests algorithm. Then, in Section 3, we use
Breiman and Cutler’s random forests FORTRAN code and the randomForest R package to
motivate our investigations into the potential issues that can emerge when the absent levels
problem is overlooked. And although a comprehensive theoretical analysis of the absent
levels problem is beyond the scope of this paper, in Section 4, we consider some simple
heuristics which may be able to help mitigate its effects. Afterwards, in Section 5, we
present three real data examples that demonstrate how the treatment of absent levels can
significantly influence a model’s performance in practice. Finally, we offer some concluding
remarks in Section 6.
2. Background
In this section, we introduce some notation and provide an overview of the random forests
algorithm. Consequently, the more knowledgeable reader may only need to review Sec-
tions 2.1.1 and 2.1.2 which cover how the algorithm’s node splits are determined.
2.1 Classification and Regression Trees (CART)
We begin by discussing the Classification and Regression Trees (CART) methodology since
the random forests algorithm uses a slightly modified version of CART to construct the
1
Breiman and Cutler’s random forests FORTRAN code is available online at:
https://www.stat.berkeley.edu/
~
breiman/RandomForests/
2
The “Absent Levels” Problem
individual decision trees that are used in its ensemble. For a more complete overview of
CART, we refer the reader to Breiman et al. (1984) or Hastie et al. (2009).
Suppose that we have a training set with N independent observations
(x
n
, y
n
) , n = 1, 2, . . . , N,
where x
n
= (x
n1
, x
n2
, . . . , x
nP
) and y
n
denote, respectively, the P -dimensional feature vector
and response for observation n. Given this initial training set, CART is a greedy recursive
binary partitioning algorithm that repeatedly partitions a larger subset of the training set
N
M
⊆ {1, 2, . . . , N} (the “mother node”) into two smaller subsets N
L
and N
R
(the “left”
and “right” daughter nodes, respectively). Each iteration of this splitting process, which
can be referred to as “growing the tree,” is accomplished by determining a decision rule that
is characterized by a “splitting variable” p ∈ {1, 2, . . . , P } and an accompanying “splitting
criterion” set S
p
which defines the subset of predictor p’s domain that gets sent to the left
daughter node N
L
. In particular, any splitting variable and splitting criterion pair (p, S
p
)
will partition the mother node N
M
into the left and right daughter nodes which are defined,
respectively, as
N
L
(p, S
p
) = {n ∈ N
M
: x
np
∈ S
p
} and N
R
(p, S
p
) =
n ∈ N
M
: x
np
∈ S
0
p
, (1)
where S
0
p
denotes the complement of the splitting criterion set S
p
with respect to predictor
p’s domain. A simple model useful for making predictions and inferences is then subse-
quently fit to the subset of the training data that is in each node.
This recursive binary partitioning procedure is continued until some stopping rule is
reached—a tuning parameter that can be controlled, for example, by placing a constraint on
the minimum number of training observations that are required in each node. Afterwards,
to help guard against overfitting, the tree can then be “pruned”—although we will not
discuss this further as pruning has not traditionally been done in the trees that are grown
in random forests (Breiman, 2001). Predictions and inferences can then be made on an
observation by first sending it down the tree according to the tree’s set of decision rules,
and then by considering the model that was fit in the furthest node of the tree that the
observation is able to reach.
The CART algorithm will grow a tree by selecting, from amongst all possible splitting
variable and splitting criterion pairs (p, S
p
), the optimal pair
p
∗
, S
∗
p
∗
which minimizes
some measure of “node impurity” in the resulting left and right daughter nodes as defined
in (1). However, the specific node impurity measure that is being minimized will depend
on whether the tree is being used for regression or classification.
In a regression tree, the responses in a node N are modeled using a constant which,
under a squared error loss, is estimated by the mean of the training responses that are in
the node—a quantity which we denote as:
ˆc(N ) = ave(y
n
| n ∈ N ) . (2)
Therefore, the CART algorithm will grow a regression tree by partitioning a mother node
N
M
on the splitting variable and splitting criterion pair
p
∗
, S
∗
p
∗
which minimizes the
squared error resulting from the two daughter nodes that are created with respect to a
3

Au
(p, S
p
) pair:
p
∗
, S
∗
p
∗
= arg min
(p,S
p
)
X
n∈N
L
(p,S
p
)
[y
n
− ˆc(N
L
(p, S
p
))]
2
+
X
n∈N
R
(p,S
p
)
[y
n
− ˆc(N
R
(p, S
p
))]
2
, (3)
where the nodes N
L
(p, S
p
) and N
R
(p, S
p
) are as defined in (1).
Meanwhile, in a classification tree where the response is categorical with K possible
response classes which are indexed by the set K = {1, 2, . . . , K}, we denote the proportion
of training observations that are in a node N belonging to each response class k as:
ˆπ
k
(N ) =
1
|N |
X
n∈N
I(y
n
= k), k ∈ K,
where |·| is the set cardinality function and I(·) is the indicator function. Node N will then
classify its observations to the majority response class
ˆ
k(N ) = arg max
k∈K
ˆπ
k
(N ) , (4)
with the Gini index
G(N ) =
K
X
k=1
[ˆπ
k
(N ) · (1 − ˆπ
k
(N ))]
providing one popular way of quantifying the node impurity in N . Consequently, the
CART algorithm will grow a classification tree by partitioning a mother node N
M
on the
splitting variable and splitting criterion pair
p
∗
, S
∗
p
∗
which minimizes the weighted Gini
index resulting from the two daughter nodes that are created with respect to a (p, S
p
) pair:
p
∗
, S
∗
p
∗
= arg min
(p,S
p
)
|N
L
(p, S
p
)| · G(N
L
(p, S
p
)) + |N
R
(p, S
p
)| · G(N
R
(p, S
p
))
|N
L
(p, S
p
)| + |N
R
(p, S
p
)|
, (5)
where the nodes N
L
(p, S
p
) and N
R
(p, S
p
) are as defined in (1).
Therefore, the CART algorithm will grow both regression and classification trees by
partitioning a mother node N
M
on the splitting variable and splitting criterion pair (p
∗
, S
∗
p
∗
)
which minimizes the requisite node impurity measure across all possible (p, S
p
) pairs—a task
which can be accomplished by first determining the optimal splitting criterion S
∗
p
for every
predictor p ∈ {1, 2, . . . , P }. However, the specific manner in which any particular predictor
p’s optimal splitting criterion S
∗
p
is determined will depend on whether p is an ordered or
categorical predictor.
2.1.1 Splitting on an Ordered Predictor
The splitting criterion S
p
for an ordered predictor p is characterized by a numeric “split
point” s
p
∈ R that defines the half-line S
p
= {x ∈ R : x ≤ s
p
}. Thus, as can be observed
from (1), a (p, S
p
) pair will partition a mother node N
M
into the left and right daughter
nodes that are defined, respectively, by
N
L
(p, S
p
) = {n ∈ N
M
: x
np
≤ s
p
} and N
R
(p, S
p
) = {n ∈ N
M
: x
np
> s
p
} .
4
The “Absent Levels” Problem
Therefore, determining the optimal splitting criterion S
∗
p
=
x ∈ R : x ≤ s
∗
p
for an ordered
predictor p is straightforward—it can be greedily found by searching through all of the
observed training values in the mother node in order to find the optimal numeric split
point s
∗
p
∈ {x
np
∈ R : n ∈ N
M
} that minimizes the requisite node impurity measure which
is given by either (3) or (5).
2.1.2 Splitting on a Categorical Predictor
For a categorical predictor p with Q possible unordered levels which are indexed by the set
Q = {1, 2, . . . , Q}, the splitting criterion S
p
⊂ Q is defined by the subset of levels that gets
sent to the left daughter node N
L
, while the complement set S
0
p
= Q \ S
p
defines the subset
of levels that gets sent to the right daughter node N
R
. For notational simplicity and ease of
exposition, in the remainder of this section we assume that all Q unordered levels of p are
present in the mother node N
M
during training since it is only these present levels which
contribute to the measure of node impurity when determining p’s optimal splitting criterion
S
∗
p
. Later, in Section 3, we extend our notation to also account for any unordered levels of
a categorical predictor p which are absent from the mother node N
M
during training.
Consequently, there are are 2
Q−1
−1 non-redundant ways of partitioning the Q unordered
levels of p into the two daughter nodes, making it computationally expensive to evaluate
the resulting measure of node impurity for every possible split when Q is large. However,
this computation simplifies in certain situations.
In the case of a regression tree with a squared error node impurity measure, a categorical
predictor p’s optimal splitting criterion S
∗
p
can be determined by using a procedure described
in Fisher (1958). Specifically, the training observations in the mother node are first used to
calculate the mean response within each of p’s unordered levels:
γ
p
(q) = ave(y
n
| n ∈ N
M
and x
np
= q) , q ∈ Q. (6)
These means are then used to assign numeric “pseudo values” ˜x
np
∈ R to every training
observation that is in the mother node according to its observed level for predictor p:
˜x
np
= γ
p
(x
np
), n ∈ N
M
. (7)
Finally, the optimal splitting criterion S
∗
p
for the categorical predictor p is determined
by doing an ordered split on these numeric pseudo values ˜x
np
—that is, a corresponding
optimal “pseudo splitting criterion”
˜
S
∗
p
=
˜x ∈ R : ˜x ≤ ˜s
∗
p
is greedily chosen by scanning
through all of the assigned numeric pseudo values in the mother node in order to find
the optimal numeric “pseudo split point” ˜s
∗
p
∈ {˜x
np
∈ R : n ∈ N
M
} which minimizes the
resulting squared error node impurity measure given in (3) with respect to the left and right
daughter nodes that are defined, respectively, by
N
L
(p,
˜
S
∗
p
) =
n ∈ N
M
: ˜x
np
≤ ˜s
∗
p
and N
R
(p,
˜
S
∗
p
) =
n ∈ N
M
: ˜x
np
> ˜s
∗
p
. (8)
Meanwhile, in the case of a classification tree with a weighted Gini index node impurity
measure, whether the computation simplifies or not is dependent on the number of response
classes. For the K > 2 multiclass classification context, no such simplification is possible,
although several approximations have been proposed (Loh and Vanichsetakul, 1988). How-
ever, for the K = 2 binary classification situation, a similar procedure to the one that was
5

Au
just described for regression trees can be used. Specifically, the proportion of the training
observations in the mother node that belong to the k = 1 response class is first calculated
within each of categorical predictor p’s unordered levels:
γ
p
(q) =
|{n ∈ N
M
: x
np
= q and y
n
= 1}|
|{n ∈ N
M
: x
np
= q}|
, q ∈ Q, (9)
and where we note here that γ
p
(q) ≥ 0 for all q since these proportions are, by definition,
nonnegative. Afterwards, and just as in equation (7), these k = 1 response class proportions
are used to assign numeric pseudo values ˜x
np
∈ R to every training observation that is in the
mother node N
M
according to its observed level for predictor p. And once again, the optimal
splitting criterion S
∗
p
for the categorical predictor p is then determined by performing an
ordered split on these numeric pseudo values ˜x
np
—that is, a corresponding optimal pseudo
splitting criterion
˜
S
∗
p
=
x ∈ R : x ≤ ˜s
∗
p
is greedily found by searching through all of the
assigned numeric pseudo values in the mother node in order to find the optimal numeric
pseudo split point ˜s
∗
p
∈ {˜x
np
∈ R : n ∈ N
M
} which minimizes the weighted Gini index node
impurity measure given by (5) with respect to the resulting two daughter nodes as defined
in (8). The proof that this procedure gives the optimal split in a binary classification tree
in terms of the weighted Gini index amongst all possible splits can be found in Breiman
et al. (1984) and Ripley (1996).
Therefore, in both regression and binary classification trees, we note that the optimal
splitting criterion S
∗
p
for a categorical predictor p can be expressed in terms of the criterion’s
associated optimal numeric pseudo split point ˜s
∗
p
and the requisite means or k = 1 response
class proportions γ
p
(q) of the unordered levels q ∈ Q of p as follows:
• The unordered levels of p that are being sent left have means or k = 1 response class
proportions γ
p
(q) that are less than or equal to ˜s
∗
p
:
S
∗
p
=
q ∈ Q : γ
p
(q) ≤ ˜s
∗
p
. (10)
• The unordered levels of p that are being sent right have means or k = 1 response class
proportions γ
p
(q) that are greater than ˜s
∗
p
:
S
∗
p
0
=
q ∈ Q : γ
p
(q) > ˜s
∗
p
. (11)
As we later discuss in Section 3, equations (10) and (11) lead to inherent differences in
the left and right daughter nodes when splitting a mother node on a categorical predictor
in CART—differences that can have significant ramifications when making predictions and
inferences for observations with absent levels.
2.2 Random Forests
Introduced in Breiman (2001), random forests are an ensemble learning method that cor-
rects for each individual tree’s propensity to overfit the training set. This is accomplished
through the use of bagging and a CART-like tree learning algorithm in order to build a
large collection of “de-correlated” decision trees.
6

The “Absent Levels” Problem
2.2.1 Bagging
Proposed in Breiman (1996a), bagging is an ensembling technique for improving the accu-
racy and stability of models. Specifically, given a training set
Z = {(x
1
, y
1
), (x
2
, y
2
), . . . , (x
N
, y
N
)} ,
this is achieved by repeatedly sampling N
0
observations with replacement from Z in order to
generate B bootstrapped training sets Z
1
, Z
2
, . . . , Z
B
, where usually N
0
= N . A separate
model is then trained on each bootstrapped training set Z
b
, where we denote model b’s
prediction on an observation x as
ˆ
f
b
(x). Here, showing each model a different bootstrapped
sample helps to de-correlate them, and the overall bagged estimate
ˆ
f(x) for an observation
x can then be obtained by averaging over all of the individual predictions in the case of
regression
ˆ
f(x) =
1
B
B
X
b=1
ˆ
f
b
(x),
or by taking the majority vote in the case of classification
ˆ
f(x) = arg max
k∈K
B
X
b=1
I
ˆ
f
b
(x) = k
!
.
One important aspect of bagging is the fact that each training observation n will only
appear “in-bag” in a subset of the bootstrapped training sets Z
b
. Therefore, for each training
observation n, an “out-of-bag” (OOB) prediction can be constructed by only considering
the subset of models in which n did not appear in the bootstrapped training set. Moreover,
an OOB error for a bagged model can be obtained by evaluating the OOB predictions for
all N training observations—a performance metric which helps to alleviate the need for
cross-validation or a separate test set (Breiman, 1996b).
2.2.2 CART-Like Tree Learning Algorithm
In the case of random forests, the model that is being trained on each individual boot-
strapped training set Z
b
is a decision tree which is grown using the CART methodology,
but with two key modifications.
First, as mentioned previously in Section 2, the trees that are grown in random forests are
generally not pruned (Breiman, 2001). And second, instead of considering all P predictors
at a split, only a randomly selected subset of the P predictors is allowed to be used—a
restriction which helps to de-correlate the trees by placing a constraint on how similarly
they can be grown. This process, which is known as the random subspace method, was
developed in Amit and Geman (1997) and Ho (1998).
3. The Absent Levels Problem
In Section 1, we defined the absent levels problem as the inherent issue for decision tree based
methods occurring whenever there is an indeterminacy over how to handle an observation
that has reached a categorical split which was determined when the observation in question’s
7
Au
level was absent during training, and we described the three different ways in which the
absent levels problem can arise. Then, in Section 2.1.2, we discussed how the levels of a
categorical predictor p which were present in the mother node N
M
during training were
used to determine its optimal splitting criterion S
∗
p
. In this section, we investigate the
potential consequences of overlooking the absent levels problem where, for a categorical
predictor p with Q unordered levels which are indexed by the set Q = {1, 2, . . . , Q}, we now
also further denote the subset of the levels of p that were present or absent in the mother
node N
M
during training, respectively, as follows:
Q
P
= {q ∈ Q : |{n ∈ N
M
: x
np
= q}| > 0} ,
Q
A
= {q ∈ Q : |{n ∈ N
M
: x
np
= q}| = 0} .
(12)
Specifically, by documenting how absent levels have been handled by Breiman and Cutler’s
random forests FORTRAN code and the randomForest R package, we show how failing to
account for the absent levels problem can systematically bias a model in practice. However,
although our investigations are motivated by these two particular software implementations
of random forests, we emphasize that the absent levels problem is, first and foremost, an
intrinsic methodological issue for decision tree based methods.
3.1 Regression
For regression trees using a squared error node impurity measure, recall from our discussions
in Section 2.1.2 and equations (6), (10), and (11), that the split of a mother node N
M
on a
categorical predictor p can be characterized in terms of the splitting criterion’s associated
optimal numeric pseudo split point ˜s
∗
p
and the means γ
p
(q) of the unordered levels q ∈ Q
of p as follows:
• The unordered levels of p being sent left have means γ
p
(q) that are less than or equal
to ˜s
∗
p
.
• The unordered levels of p being sent right have means γ
p
(q) that are greater than ˜s
∗
p
.
Furthermore, recall from (2), that a node’s prediction is given by the mean of the training
responses that are in the node. Therefore, because the prediction of each daughter node can
be expressed as a weighted average over the means γ
p
(q) of the present levels q ∈ Q
P
that
are being sent to it, it follows that the left daughter node N
L
will always give a prediction
that is smaller than the right daughter node N
R
when splitting on a categorical predictor p
in a regression tree that uses a squared error node impurity measure.
In terms of execution, both the random forests FORTRAN code and the randomForest R
package employ the pseudo value procedure for regression that was described in Section 2.1.2
when determining the optimal splitting criterion S
∗
p
for a categorical predictor p. However,
the code that is responsible for calculating the mean γ
p
(q) within each unordered level q ∈ Q
as in equation (6) behaves as follows:
γ
p
(q) =
(
ave(y
n
| n ∈ N
M
and x
np
= q) if q ∈ Q
P
0 if q ∈ Q
A
,
where Q
P
and Q
A
are, respectively, the present and absent levels of p as defined in (12).
8
The “Absent Levels” Problem
Although this “zero imputation” of the means γ
p
(q) for the absent levels q ∈ Q
A
is
inconsequential when determining the optimal numeric pseudo split point ˜s
∗
p
during training,
it can be highly influential on the subsequent predictions that are made for observations
with absent levels. In particular, by (10) and (11), the absent levels q ∈ Q
A
will be sent left
if ˜s
∗
p
≥ 0, and they will be sent right if ˜s
∗
p
< 0. But, due to the systematic differences that
exist amongst the two daughter nodes, this arbitrary decision of sending the absent levels
left versus right can significantly impact the predictions that are made on observations with
absent levels—even though the model’s final predictions will also depend on any ensuing
splits which take place after the absent levels problem occurs, observations with absent
levels will tend to be biased towards smaller predictions when they are sent to the left
daughter node, and they will tend to be biased towards larger predictions when they are
sent to the right daughter node.
In addition, this behavior also implies that the random forest regression models which
are trained using either the random forests FORTRAN code or the randomForest R package
are sensitive to the set of possible values that the training responses can take. To illustrate,
consider the following two extreme cases when splitting a mother node N
M
on a categorical
predictor p:
• If the training responses y
n
> 0 for all n, then the pseudo numeric split point ˜s
∗
p
> 0
since the means γ
p
(q) > 0 for all of the present levels q ∈ Q
P
. And because the
“imputed” means γ
p
(q) = 0 < ˜s
∗
p
for all q ∈ Q
A
, the absent levels will always be sent
to the left daughter node N
L
which gives smaller predictions.
• If the training responses y
n
< 0 for all n, then the pseudo numeric split point ˜s
∗
p
< 0
since the means γ
p
(q) < 0 for all of the present levels q ∈ Q
P
. And because the
“imputed” means γ
p
(q) = 0 > ˜s
∗
p
for all q ∈ Q
A
, the absent levels will always be sent
to the right daughter node N
R
which gives larger predictions.
And although this sensitivity to the training response values was most easily demonstrated
through these two extreme situations, the reader should not let this overshadow the fact
that the absent levels problem can also heavily influence a model’s performance in more
general circumstances (e.g., when the training responses are of mixed signs).
3.2 Classification
For binary classification trees using a weighted Gini index node impurity measure, recall
from our discussions in Section 2.1.2 and equations (9), (10), and (11), that the split of a
mother node N
M
on a categorical predictor p can be characterized in terms of the splitting
criterion’s associated optimal numeric pseudo split point ˜s
∗
p
and the k = 1 response class
proportions γ
p
(q) of the unordered levels q ∈ Q of p as follows:
• The unordered levels of p being sent left have k = 1 response class proportions γ
p
(q)
that are less than or equal to ˜s
∗
p
.
• The unordered levels of p being sent right have k = 1 response class proportions γ
p
(q)
that are greater than ˜s
∗
p
.
In addition, recall from (4), that a node’s classification is given by the response class that
occurs the most amongst the training observations that are in the node. Therefore, because
9

Au
the response class proportions of each daughter node can be expressed as a weighted average
over the response class proportions of the present levels q ∈ Q
P
that are being sent to it,
it follows that the left daughter node N
L
is always less likely to classify an observation to
the k = 1 response class than the right daughter node N
R
when splitting on a categorical
predictor p in a binary classification tree that uses a weighted Gini index node impurity
measure.
In terms of implementation, the randomForest R package uses the pseudo value pro-
cedure for binary classification that was described in Section 2.1.2 when determining the
optimal splitting criterion S
∗
p
for a categorical predictor p with a “large” number of un-
ordered levels.
2
However, the code that is responsible for computing the k = 1 response
class proportion γ
p
(q) within each unordered level q ∈ Q as in equation (9) executes as
follows:
γ
p
(q) =
(
|{n ∈ N
M
: x
np
= q and y
n
= 1}|
|{n ∈ N
M
: x
np
= q}|
if q ∈ Q
P
0 if q ∈ Q
A
.
Therefore, the issues that arise here are similar to the ones that were described for regression.
Even though this “zero imputation” of the k = 1 response class proportions γ
p
(q) for the
absent levels q ∈ Q
A
is unimportant when determining the optimal numeric pseudo split
point ˜s
∗
p
during training, it can have a large effect on the subsequent classifications that are
made for observations with absent levels. In particular, since the proportions γ
p
(q) ≥ 0 for
all of the present levels q ∈ Q
P
, it follows from our discussions in Section 2.1.2 that the
numeric pseudo split point ˜s
∗
p
≥ 0. And because the “imputed” proportions γ
p
(q) = 0 ≤ ˜s
∗
p
for all q ∈ Q
A
, the absent levels will always be sent to the left daughter node. But,
due to the innate differences that exist amongst the two daughter nodes, this arbitrary
choice of sending the absent levels left can significantly affect the classifications that are
made on observations with absent levels—although the model’s final classifications will also
depend on any successive splits which take place after the absent levels problem occurs,
the classifications for observations with absent levels will tend to be biased towards the
k = 2 response class. Moreover, this behavior also implies that the random forest binary
classification models which are trained using the randomForest R package may be sensitive
to the actual ordering of the response classes: since observations with absent levels are
always sent to the left daughter node N
L
which is more likely to classify them to the k = 2
response class than the right daughter node N
R
, the classifications for these observations
can be influenced by interchanging the indices of the two response classes.
Meanwhile, for cases where the pseudo value procedure is not or cannot be used, the
random forests FORTRAN code and the randomForest R package will instead adopt a more
brute force approach that either exhaustively or randomly searches through the space of
possible splits. However, to understand the potential problems that absent levels can cause
in these situations, we must first briefly digress into a discussion of how categorical splits
are internally represented in their code.
Specifically, in their code, a split on a categorical predictor p is both encoded and
decoded as an integer whose binary representation identifies which unordered levels go left
2
The exact condition for using the pseudo value procedure for binary classification in version 4.6-12 of
the randomForest R package is when a categorical predictor p has Q > 10 unordered levels. Meanwhile,
although the random forests FORTRAN code for binary classification references the pseudo value procedure, it
does not appear to be implemented in the code.
10

The “Absent Levels” Problem
(the bits that are “turned on”) and which unordered levels go right (the bits that are
“turned off”). To illustrate, consider the situation where a categorical predictor p has four
unordered levels, and where the integer encoding of the split is 5. In this case, since 0101
is the binary representation of the integer 5 (because 5 = [0] · 2
3
+ [1] · 2
2
+ [0] · 2
1
+ [1] · 2
0
),
levels 1 and 3 get sent left while levels 2 and 4 get sent right.
Now, when executing an exhaustive search to find the optimal splitting criterion S
∗
p
for a categorical predictor p with Q unordered levels, the random forests FORTRAN code
and the randomForest R package will both follow the same systematic procedure:
3
all
2
Q−1
− 1 possible integer encodings for the non-redundant partitions of the unordered levels
of predictor p are evaluated in increasing sequential order starting from 1 and ending at
2
Q−1
− 1 , with the choice of the optimal splitting criterion S
∗
p
being updated if and only if
the resulting weighted Gini index node impurity measure strictly improves.
But since the absent levels q ∈ Q
A
are not present in the mother node N
M
during
training, sending them left or right has no effect on the resulting weighted Gini index. And
because turning on the bit for any particular level q while holding the bits for all of the
other levels constant will always result in a larger integer, it follows that the exhaustive
search that is used by these two software implementations will always prefer splits that send
all of the absent levels right since they are always checked before any of their analogous Gini
index equivalent splits that send some of the absent levels left.
Furthermore, in their exhaustive search, the leftmost bit corresponding to the Q
th
in-
dexed unordered level of a categorical predictor p is always turned off since checking the
splits where this bit is turned on would be redundant—they would amount to just swap-
ping the “left” and “right” daughter node labels for splits that have already been evaluated.
Consequently, the Q
th
indexed level of p will also always be sent to the right daughter node
and, as a result, the classifications for observations with absent levels will tend to be biased
towards the response class distribution of the training observations in the mother node N
M
that belong to this Q
th
indexed level. Therefore, although it may sound contradictory, this
also implies that the random forest multiclass classification models which are trained using
either the random forests FORTRAN code or the randomForest R package may be sensitive
to the actual ordering of a categorical predictor’s unordered levels—a reordering of these
levels could potentially interchange the “left” and “right” daughter node labels, which could
then subsequently affect the classifications that are made for observations with absent levels
since they will always be sent to whichever node ends up being designated as the “right”
daughter node.
Finally, when a categorical predictor p has too many levels for an exhaustive search to
be computationally efficient, both the random forests FORTRAN code and the randomForest
R package will resort to approximating the optimal splitting criterion S
∗
p
with the best split
that was found amongst a large number of randomly generated splits.
4
This is accomplished
by randomly setting all of the bits in the binary representations of the splits to either a
3
The random forests FORTRAN code will use an exhaustive search for both binary and multiclass classi-
fication whenever Q < 25. In version 4.6-12 of the randomForest R package, an exhaustive search will be
used for both binary and multiclass classification whenever Q < 10.
4
The random forests FORTRAN code will use a random search for both binary and multiclass classification
whenever Q ≥ 25. In version 4.6-12 of the randomForest R package, a random search will only be used when
Q ≥ 10 in the multiclass classification case.
11

Au
0 or a 1—a procedure which ultimately results in each absent level being randomly sent
to either the left or right daughter node with equal probability. As a result, although the
absent levels problem can still occur in these situations, it is difficult to determine whether
it results in any systematic bias. However, it is still an open question as to whether or not
such a treatment of absent levels is sufficient.
4. Heuristics for Mitigating the Absent Levels Problem
Although a comprehensive theoretical analysis of the absent levels problem is beyond the
scope of this paper, in this section we briefly consider several heuristics which may be able to
help mitigate the issue. Later, in Section 5, we empirically evaluate and compare how some
of these heuristics perform in practice when they are applied to three real data examples.
4.1 Missing Data Heuristics
Even though absent levels are fully observed and known, the missing data literature for
decision tree based methods is still perhaps the area of existing research that is most closely
related to the absent levels problem.
4.1.1 Stop
One straightforward missing data strategy for dealing with absent levels would be to simply
stop an observation from going further down the tree whenever the issue occurs and just
use the mother node for prediction—a missing data approach which has been adopted
by both the rpart R package for CART (Therneau et al., 2015) and the gbm R package
for generalized boosted regression models (Ridgeway, 2013). Even with this missing data
functionality already in place, however, the gbm R package has still had its own issues in
readily extending it to the case of absent levels—serving as another example of a software
implementation of a decision tree based method that has overlooked and suffered from the
absent levels problem.
5
4.1.2 Distribution-Based Imputation (DBI)
Another potential missing data technique would be to send an observation with an absent
level down both daughter nodes—perhaps by using the distribution-based imputation (DBI)
technique which is employed by the C4.5 algorithm for growing decision trees (Quinlan,
1993). In particular, an observation that encounters an infeasible node split is first split
into multiple pseudo-instances, where each instance takes on a different imputed value
and weight based on the distribution of observed values for the splitting variable in the
mother node’s subset of the training data. These pseudo-instances are then sent down
their appropriate daughter nodes in order to proceed down the tree as usual, and the
final prediction is derived from the weighted predictions of all the terminal nodes that are
subsequently reached (Saar-Tsechansky and Provost, 2007).
5
See, for example, https://code.google.com/archive/p/gradientboostedmodels/issues/7
12
The “Absent Levels” Problem
4.1.3 Surrogate Splits
Surrogate splitting, which the rpart R package also supports, is arguably the most popular
method of handling missing data in CART, and it may provide another workable approach
for mitigating the effects of absent levels. Specifically, if
p
∗
, S
∗
p
∗
is found to be the optimal
splitting variable and splitting criterion pair for a mother node N
M
, then the first surrogate
split is the (p
0
, S
p
0
) pair where p
0
6= p
∗
that yields the split which most closely mimics the
optimal split’s binary partitioning of N
M
, the second surrogate split is the (p
00
, S
p
00
) pair
where p
00
6∈ {p
∗
, p
0
} resulting in the second most similar binary partitioning of N
M
as the
optimal split, and so on. Afterwards, when an observation reaches an indeterminate split,
the surrogates are tried in the order of decreasing similarity until one of them becomes
feasible (Breiman et al., 1984).
However, despite its extensive use in CART, surrogate splitting may not be entirely
appropriate for ensemble tree methods like random forests. As pointed out in Ishwaran
et al. (2008):
Although surrogate splitting works well for trees, the method may not be well
suited for forests. Speed is one issue. Finding a surrogate split is computation-
ally intensive and may become infeasible when growing a large number of trees,
especially for fully saturated trees used by forests. Further, surrogate splits may
not even be meaningful in a forest paradigm. [Random forests] randomly selects
variables when splitting a node and, as such, variables within a node may be
uncorrelated, and a reasonable surrogate split may not exist. Another concern
is that surrogate splitting alters the interpretation of a variable, which affects
measures such as [variable importance].
Nevertheless, surrogate splitting is still available as a non-default option for handling missing
data in the partykit R package (Hothorn and Zeileis, 2015), which is an implementation of a
bagging ensemble of conditional inference trees that correct for the biased variable selection
issues which exist in several tree learning algorithms like CART and C4.5 (Hothorn et al.,
2006).
4.1.4 Random/Majority
The partykit R package also provides some other functionality for dealing with missing
data that may be applicable to the absent levels problem. These include the package’s de-
fault approach of randomly sending the observations to one of the two daughter nodes with
the weighting done by the number of training observations in each node or, alternatively,
by simply having the observations go to the daughter node with more training observa-
tions. Interestingly, the partykit R package does appear to recognize the possibility of
absent levels occurring, and chooses to handle them as if they were missing—its reference
manual states that “Factors in test samples whose levels were empty in the learning sample
are treated as missing when computing predictions.” Whether or not such missing data
heuristics adequately address the absent levels problem, however, is still unknown.
13

Au
4.2 Feature Engineering Heuristics
Apart from missing data methods, feature engineering techniques which transform the cat-
egorical predictors may also be viable approaches to mitigating the effects of absent levels.
However, feature engineering techniques are not without their own drawbacks. First,
transforming the categorical predictors may not always be feasible in practice since the fea-
ture space may become computationally unmanageable. And even when transformations are
possible, they may further exacerbate variable selection issues—many popular tree learn-
ing algorithms such as CART and C4.5 are known to be biased in favor of splitting on
ordered predictors and categorical predictors with many unordered levels since they offer
more candidate splitting points to choose from (Hothorn et al., 2006). Moreover, by recod-
ing a categorical predictor’s unordered levels into several different predictors, we forfeit a
decision tree based method’s natural ability to simultaneously consider all of the predictor’s
levels together at a single split. Thus, it is not clear whether feature engineering techniques
are preferable when using decision tree based methods.
Despite these potential shortcomings, transformations of the categorical predictors is
currently required by the scikit-learn Python module’s implementation of random forests
(Pedregosa et al., 2011). There have, however, been some discussions about extending
the module so that it can support the native categorical split capabilities used by the
random forests FORTRAN code and the randomForest R package.
6
But, needless to say, such
efforts would also have the unfortunate consequence of introducing the indeterminacy of
the absent levels problem into another popular software implementation of a decision tree
based method.
4.2.1 One-Hot Encoding
Nevertheless, one-hot encoding is perhaps the most straightforward feature engineering
technique that could be applied to the absent levels problem—even though some unordered
levels may still be absent when determining a categorical split during training, any uncer-
tainty over where to subsequently send these absent levels would be eliminated by recoding
the levels of each categorical predictor into separate dummy predictors.
5. Examples
Although the actual severity of the absent levels problem will depend on the specific data
set and task at hand, in this section we present three real data examples which illustrate
how the absent levels problem can dramatically alter the performance of decision tree based
methods in practice. In particular, we empirically evaluate and compare how the seven
different heuristics in the set
H = {Left, Right, Stop, Majority, Random, DBI, One-Hot}
perform when confronted with the absent levels problem in random forests.
In particular, the first two heuristics that we consider in our set H are the systematically
biased approaches discussed in Section 3 which have been employed by both the random
6
See, for example, https://github.com/scikit-learn/scikit-learn/pull/3346
14

The “Absent Levels” Problem
forests FORTRAN code and the randomForest R package due to having overlooked the absent
levels problem:
• Left: Sending the observation to the left daughter node.
• Right: Sending the observation to the right daughter node.
Consequently, these two “naive heuristics” have been included in our analysis for compar-
ative purposes only.
In our set of heuristics H, we also consider some of the missing data strategies for
decision tree based methods that we discussed in Section 4:
• Stop: Stopping the observation from going further down the tree and using the
mother node for prediction.
• Majority: Sending the observation to the daughter node with more training obser-
vations, with any ties being broken randomly.
• Random: Randomly sending the observation to one of the two daughter nodes, with
the weighting done by the number of training observations in each node.
7
• Distribution-Based Imputation (DBI): Sending the observation to both daughter
nodes using the C4.5 tree learning algorithm’s DBI approach.
Unlike the two naive heuristics, these “missing data heuristics” are all less systematic in
their preferences amongst the two daughter nodes.
Finally, in our set H, we also consider a “feature engineering heuristic” which transforms
all of the categorical predictors in the original data set:
• One-Hot: Recoding every categorical predictor’s set of possible unordered levels into
separate dummy predictors
Under this heuristic, although unordered levels may still be absent when determining a cat-
egorical split during training, there is no longer any uncertainty over where to subsequently
send observations with absent levels.
Code for implementing the naive and missing data heuristics was built on top of version
4.6-12 of the randomForest R package. Specifically, the randomForest R package is used
to first train the random forest models as usual. Afterwards, each individual tree’s in-bag
training data is sent back down the tree according to the tree’s set of decision rules in
order to record the unordered levels that were absent at each categorical split. Finally,
when making predictions or inferences, our code provides some functionality for carrying
out each of the naive and missing data heuristics whenever the absent levels problem occurs.
Each of the random forest models that we consider in our analysis is trained “off-the-
shelf” by using the randomForest R package’s default settings for the algorithm’s tuning
7
We also investigated an alternative “unweighted” version of the Random heuristic which randomly sends
observations with absent levels to either the left or right daughter node with equal probability (analogous
to the random search procedure that was described at the end of Section 3.2). However, because this
unweighted version was found to be generally inferior to the “weighted” version described in our analysis,
we have omitted it from our discussions for expositional clarity and conciseness.
15
Au
0
20
40
60
0.00 0.25 0.50 0.75 1.00
1985 Auto Imports
0
20
40
60
80
0.00 0.25 0.50 0.75 1.00
PROMESA
0
5
10
0.00 0.25 0.50 0.75 1.00
Pittsburgh Bridges
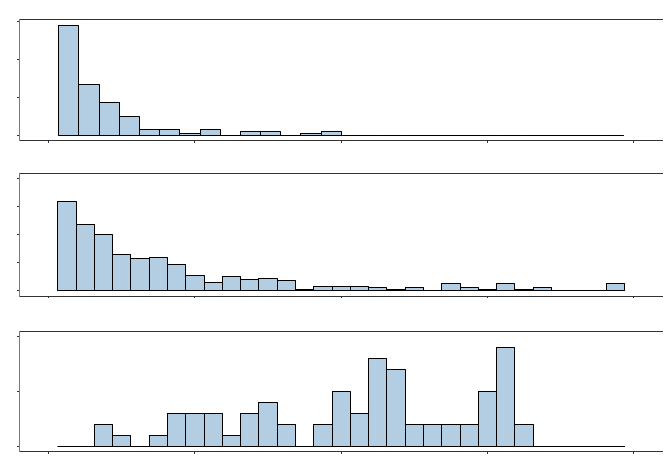
OOB Absence Proportion
Frequency
Figure 1: Histograms for each example’s distribution of OOB absence proportions.
parameters. Moreover, to account for the inherent randomness in the random forests al-
gorithm, we repeat each of our examples 1000 times with a different random seed used to
initialize each experimental replication. However, because of the way in which we have
structured our code, we note that our analysis is able to isolate the effects of the naive
and missing data heuristics on the absent levels problem since, within each experimental
replication, their underlying random forest models are identical with respect to each tree’s
in-bag training data and differ only in terms of their treatment of the absent levels. As a
result, the predictions and inferences obtained from the naive and missing data heuristics
will be positively correlated across the 1000 experimental replications that we consider for
each example—a fact which we exploit in order to improve the precision of our comparisons.
Recall from Section 4, however, that the random forest models which are trained on fea-
ture engineered data sets are intrinsically different from the random forest models which are
trained on their original untransformed data set counterparts. Therefore, although we use
the same default randomForest R package settings and the same random seed to initialize
each of the One-Hot heuristic’s experimental replications, we note that its predictions and
inferences will be essentially uncorrelated with the naive and missing data heuristics across
each example’s 1000 experimental replications.
5.1 1985 Auto Imports
For a regression example, we consider the 1985 Auto Imports data set from the UCI Machine
Learning Repository (Lichman, 2013) which, after discarding observations with missing
data, contains 25 predictors that can be used to predict the prices of 159 cars. Categorical
predictors for which the absent levels problem can occur include a car’s make (18 levels),
16

The “Absent Levels” Problem
Statistic 1985 Auto Imports PROMESA Pittsburgh Bridges
Min 0.003 0.001 0.080
1st Quartile 0.021 0.020 0.368
Median 0.043 0.088 0.564
Mean 0.076 0.162 0.526
3rd Quartile 0.093 0.206 0.702
Max 0.498 0.992 0.820
Table 1: Summary statistics for each example’s distribution of OOB absence proportions.
body style (5 levels), drive layout (3 levels), engine type (5 levels), and fuel system (6 levels).
Furthermore, because all of the car prices are positive, we know from Section 3.1 that the
random forests FORTRAN code and the randomForest R package will both always employ the
Left heuristic when faced with absent levels for this particular data set.
8
The top panel in Figure 1 depicts a histogram of this example’s OOB absence propor-
tions, which we define for each training observation as the proportion of its OOB trees
across all 1000 experimental replications which had the absent levels problem occur at least
once when using the training set with the original untransformed categorical predictors.
Meanwhile, Table 1 provides a more detailed summary of this example’s distribution of
OOB absence proportions. Consequently, although there is a noticeable right skew in the
distribution, we see that most of the observations in this example had the absent levels
problem occur in less than 5% of their OOB trees.
Let ˆy
(h)
nr
denote the OOB prediction that a heuristic h makes for an observation n
in an experimental replication r. Then, within each experimental replication r, we can
compare the predictions that two different heuristics h
1
, h
2
∈ H make for an observation
n by considering the difference ˆy
(h
1
)
nr
− ˆy
(h
2
)
nr
. We summarize these comparisons for all
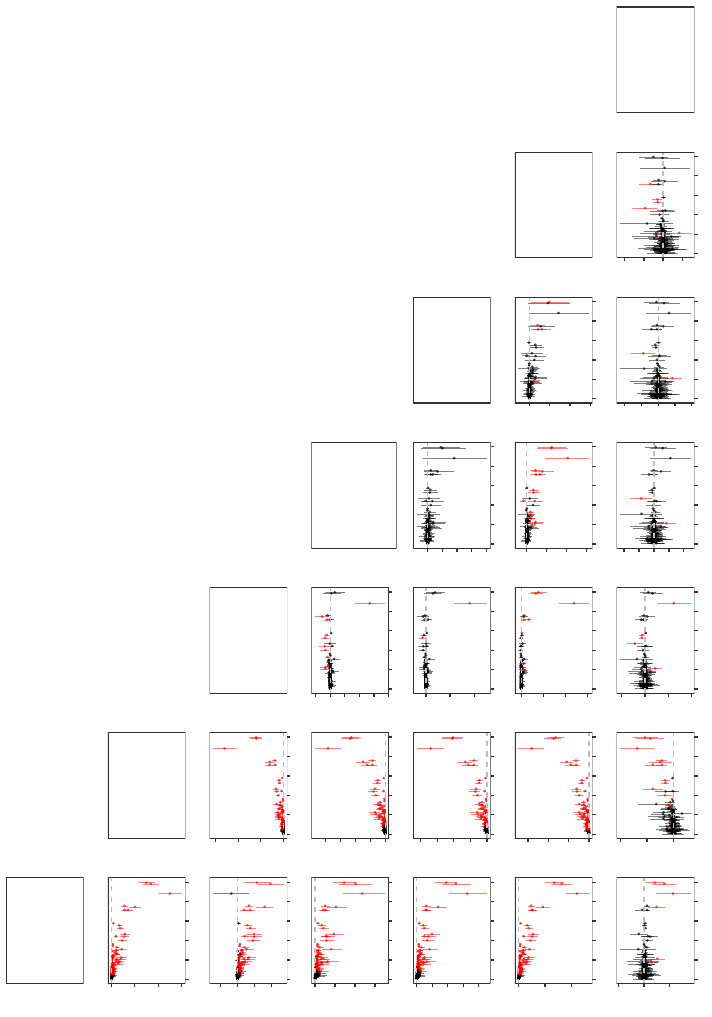
possible pairwise combinations of the seven heuristics in Figure 2, where each panel plots
the mean and middle 95% of these differences across all 1000 experimental replications as
a function of the OOB absence proportion. From the red intervals in Figure 2, we see
that significant differences in the predictions of the heuristics do exist, with the magnitude
of the point estimates and the width of the intervals tending to increase with the OOB
absence proportion—behavior that agrees with our intuition that the distinctive effects of
each heuristic should become more pronounced the more often the absent levels problem
occurs.
In addition, we can evaluate the overall performance of each heuristic h ∈ H within an
experimental replication r in terms of its root mean squared error (RMSE):
RMSE
(h)
r
=
v
u
u
t
1
N
N
X
n=1
y
n
− ˆy
(h)
nr
2
.
8
This is the case for versions 4.6-7 and earlier of the randomForest R package. Beginning in version 4.6-9,
however, the randomForest R package began to internally mean center the training responses prior to fitting
the model, with the mean being subsequently added back to the predictions of each node. Consequently, the
Left heuristic isn’t always used in these versions of the randomForest R package since the training responses
that the model actually considers are of mixed sign. Nevertheless, such a strategy still fails to explicitly
address the underlying absent levels problem.
17
Au
Left
−6000
−4000
−2000
0
0.0 0.1 0.2 0.3 0.4 0.5
Right
−1000
−500
0
500
0.0 0.1 0.2 0.3 0.4 0.5
0
2000
4000
6000
0.0 0.1 0.2 0.3 0.4 0.5
Stop
−1500
−1000
−500
0
0.0 0.1 0.2 0.3 0.4 0.5
0
1000
2000
3000
4000
0.0 0.1 0.2 0.3 0.4 0.5
−2000
−1500
−1000
−500
0
500
0.0 0.1 0.2 0.3 0.4 0.5
Majority
−2000
−1500
−1000
−500
0
0.0 0.1 0.2 0.3 0.4 0.5
0
1000
2000
3000
4000
0.0 0.1 0.2 0.3 0.4 0.5
−2000
−1000
0
0.0 0.1 0.2 0.3 0.4 0.5
−1000
−750
−500
−250
0
0.0 0.1 0.2 0.3 0.4 0.5
Random
−2000
−1000
0
0.0 0.1 0.2 0.3 0.4 0.5
0
1000
2000
3000
0.0 0.1 0.2 0.3 0.4 0.5
−3000
−2000
−1000
0
0.0 0.1 0.2 0.3 0.4 0.5
−1500
−1000
−500
0
0.0 0.1 0.2 0.3 0.4 0.5
−1200
−800
−400
0
0.0 0.1 0.2 0.3 0.4 0.5
DBI
−2000
0
2000
0.0 0.1 0.2 0.3 0.4 0.5
0
2000
4000
0.0 0.1 0.2 0.3 0.4 0.5
−4000
−2000
0
2000
0.0 0.1 0.2 0.3 0.4 0.5
−2000
−1000
0
1000
2000
0.0 0.1 0.2 0.3 0.4 0.5
−2000
−1000
0
1000
2000
0.0 0.1 0.2 0.3 0.4 0.5
−1000
0
1000
2000
0.0 0.1 0.2 0.3 0.4 0.5
One−Hot
OOB Absence Proportion
Difference in OOB Prediction
Figure 2: Pairwise differences in the OOB predictions as a function of the OOB absence proportions in the 1985 Auto Imports
data set. Each panel plots the mean and middle 95% of the differences across all 1000 experimental replications when the
OOB predictions of the heuristic that is labeled at the right of the panel’s row is subtracted from the OOB predictions
of the heuristic that is labeled at the top of the panel’s column. Differences were taken within each experimental
replication in order to account for the positive correlation that exists between the naive and missing data heuristics.
Intervals containing zero (the horizontal dashed line) are in black, while intervals not containing zero are in red.
18
The “Absent Levels” Problem
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
1900
2000
2100
2200
Left Right Stop Majority Random DBI One−Hot
RMSE
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
−5
0
5
Left Right Stop Majority Random DBI One−Hot
Relative RMSE (%)
Absent Levels Heuristic
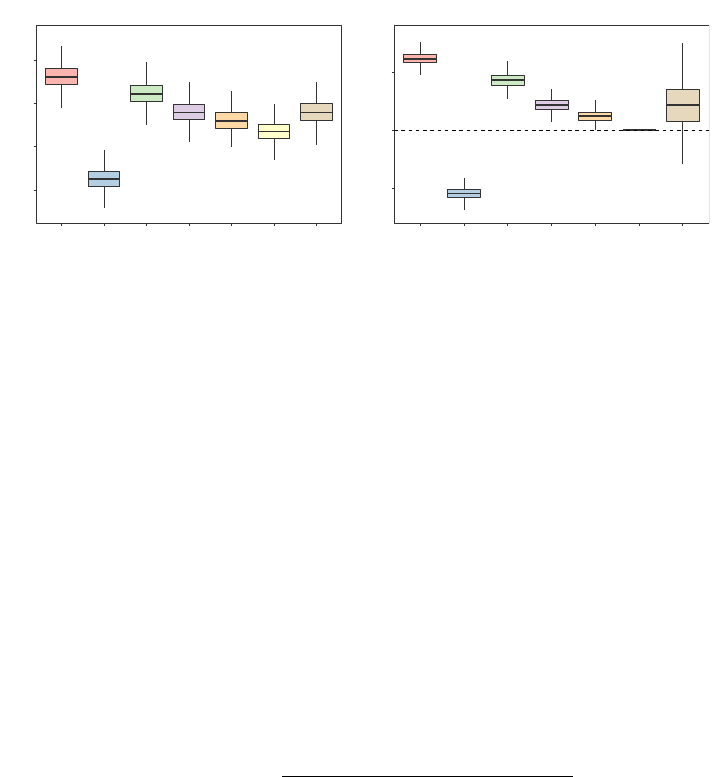
Figure 3: RMSEs for the OOB predictions of the seven heuristics in the 1985 Auto Imports
data set. The left panel shows boxplots of each heuristic’s marginal distribution of
RMSEs across all 1000 experimental replications, which ignores the positive corre-
lation that exists between the naive and missing data heuristics. The right panel
accounts for this positive correlation by comparing the RMSEs of the heuristics
relative to the best RMSE that was obtained amongst the missing data heuristics
within each of the 1000 experimental replications as in (13).
Boxplots displaying each heuristic’s marginal distribution of RMSEs across all 1000 ex-
perimental replications are shown in the left panel of Figure 3. However, these marginal
boxplots ignore the positive correlation that exists between the naive and missing data
heuristics. Therefore, within every experimental replication r, we also compare the RMSE
for each heuristic h ∈ H relative to the best RMSE that was achieved amongst the missing
data heuristics H
m
= {Stop, Majority, Random, DBI }:
RMSE
(hkH
m
)
r
=
RMSE
(h)
r
− min
h∈H
m
RMSE
(h)
r
min
h∈H
m
RMSE
(h)
r
. (13)
Here we note that the Left and Right heuristics were not considered in the definition of the
best RMSE achieved within each experimental replication r due to the issues discussed in
Section 3, while the One-Hot heuristic was excluded from this definition since it is essentially
uncorrelated with the other six heuristics across all 1000 experimental replications. Boxplots
of these relative RMSEs are shown in the right panel of Figure 3.
5.1.1 Naive Heuristics
Relative to all of the other heuristics and consistent with our discussions in Section 3.1, we
see from Figure 2 that the Left and Right heuristics have a tendency to severely underpredict
and overpredict, respectively. Furthermore, for this particular example, we notice from
Figure 3 that the random forests FORTRAN code and the randomForest R package’s behavior
of always sending absent levels left in this particular data set substantially underperforms
relative to the other heuristics—it gives an RMSE that is, on average, 6.2% worse than
the best performing missing data heuristic. And although the Right heuristic appears to
19
Au
perform exceptionally well, we again stress the misleading nature of this performance—its
tendency to overpredict just coincidentally happens to be beneficial in this specific situation.
5.1.2 Missing Data Heuristics
As can be seen from Figure 2, the predictions obtained from the four missing data heuristics
are more aligned with one another than they are with the Left, Right, and One-Hot heuris-
tics. Considerable disparities in their predictions do still exist, however, and from Figure 3
we note that amongst the four missing data heuristics, the DBI heuristic clearly performs the
best. And although the Majority heuristic fares slightly worse than the Random heuristic,
they both perform appreciably better than the Stop heuristic.
5.1.3 Feature Engineering Heuristic
Recall that the One-Hot heuristic is essentially uncorrelated with the other six heuristics
across all 1000 experimental replications—a fact which is reflected in its noticeably wider
intervals in Figure 2 and in its larger relative RMSE boxplot in Figure 3. Nevertheless, it
can still be observed from Figure 3 that although the One-Hot heuristic’s predictions are
sometimes able to outperform the other heuristics, on average, it yields an RMSE that is
2.2% worse than than the best performing missing data heuristic.
5.2 PROMESA
For a binary classification example, we consider the June 9, 2016 United States House of
Representatives vote on the Puerto Rico Oversight, Management, and Economic Stability
Act (PROMESA) for addressing the Puerto Rican government’s debt crisis. Data for this
vote was obtained by using the Rvoteview R package to query the Voteview database (Lewis,
2015). After omitting those who did not vote on the bill, the data set contains four predictors
that can be used to predict the binary “No” or “Yes” votes of 424 House of Representative
members. These predictors include a categorical predictor for a representative’s political
party (2 levels), a categorical predictor for a representative’s state (50 levels), and two
ordered predictors which quantify aspects of a representative’s political ideological position
(McCarty et al., 1997).
The “No” vote was taken to be the k = 1 response class in our analysis, while the “Yes”
vote was taken to be the k = 2 response class. Recall from Section 3.2, that this ordering of
the response classes is meaningful in a binary classification context since the randomForest
R package will always use the Left heuristic which biases predictions for observations with
absent levels towards whichever response class is indexed by k = 2 (corresponding to the
“Yes” vote in our analysis).
From Figure 1 and Table 1, we see that the absent levels problem occurs much more
frequently in this example than it did in our 1985 Auto Imports example. In particular, the
seven House of Representative members who were the sole representatives from their state
had OOB absence proportions that were greater than 0.961 since the absent levels problem
occurred for these observations every time they reached an OOB tree node that was split
on the state predictor.
For random forest classification models, the predicted probability that an observation
belongs to a response class k can be estimated by the proportion of the observation’s trees
20
The “Absent Levels” Problem
Left
0.00
0.25
0.50
0.75
1.00
0.00 0.25 0.50 0.75 1.00
Right
−0.1
0.0
0.1
0.2
0.3
0.4
0.00 0.25 0.50 0.75 1.00
−0.75
−0.50
−0.25
0.00
0.00 0.25 0.50 0.75 1.00
Stop
0.0
0.2
0.4
0.6
0.00 0.25 0.50 0.75 1.00
−0.6
−0.4
−0.2
0.0
0.00 0.25 0.50 0.75 1.00
0.0
0.2
0.4
0.00 0.25 0.50 0.75 1.00
Majority
0.0
0.2
0.4
0.00 0.25 0.50 0.75 1.00
−0.6
−0.4
−0.2
0.0
0.00 0.25 0.50 0.75 1.00
0.0
0.2
0.4
0.00 0.25 0.50 0.75 1.00
−0.1
0.0
0.1
0.00 0.25 0.50 0.75 1.00
Random
0.0
0.2
0.4
0.00 0.25 0.50 0.75 1.00
−0.8
−0.6
−0.4
−0.2
0.0
0.00 0.25 0.50 0.75 1.00
−0.1
0.0
0.1
0.2
0.3
0.00 0.25 0.50 0.75 1.00
−0.2
−0.1
0.0
0.00 0.25 0.50 0.75 1.00
−0.2
−0.1
0.0
0.1
0.00 0.25 0.50 0.75 1.00
DBI
−0.50
−0.25
0.00
0.25
0.50
0.00 0.25 0.50 0.75 1.00
−0.75
−0.50
−0.25
0.00
0.25
0.00 0.25 0.50 0.75 1.00
−0.50
−0.25
0.00
0.25
0.00 0.25 0.50 0.75 1.00
−0.50
−0.25
0.00
0.25
0.00 0.25 0.50 0.75 1.00
−0.4
−0.2
0.0
0.2
0.00 0.25 0.50 0.75 1.00
−0.50
−0.25
0.00
0.25
0.00 0.25 0.50 0.75 1.00
One−Hot
OOB Absence Proportion
Difference in OOB Prediction
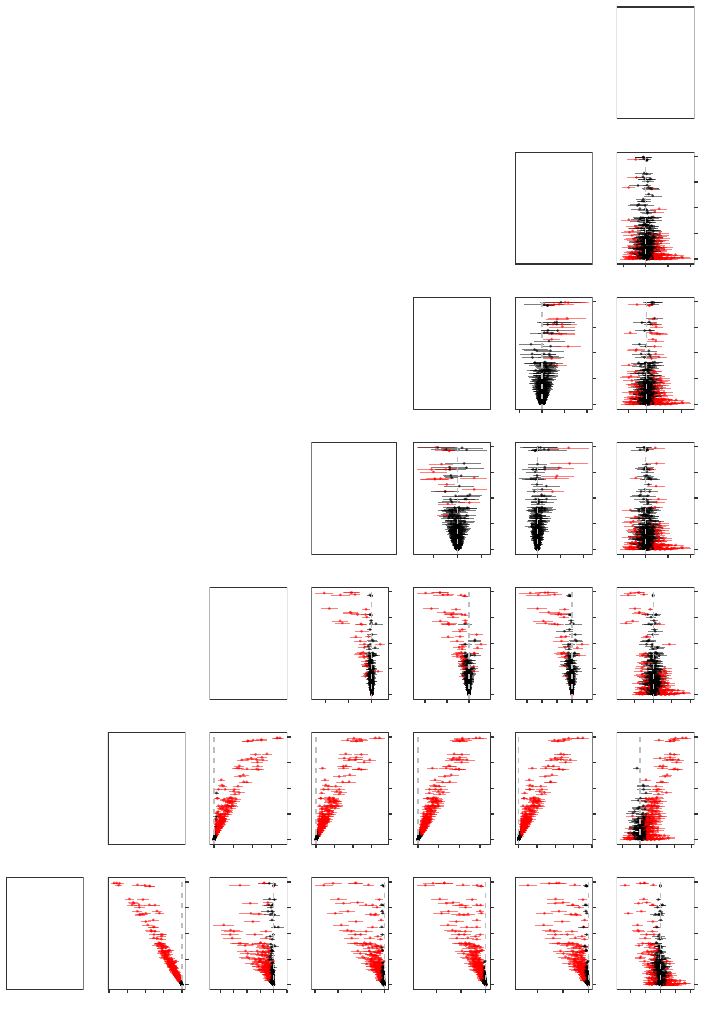
Figure 4: Pairwise differences in the OOB predicted probabilities of voting “Yes” as a function of the OOB absence proportion
in the PROMESA data set. Each panel plots the mean and middle 95% of the pairwise differences across all 1000
experimental replications when the OOB predicted probabilities of the heuristic that is labeled at the right of the
panel’s row is subtracted from the OOB predicted probabilities of the heuristic that is labeled at the top of the panel’s
column. Differences were taken within each experimental replication to account for the positive correlation that exists
between the naive and missing data heuristics. Intervals containing zero (the horizontal dashed line) are in black, while
intervals not containing zero are in red.
21

Au
which classify it to class k.
9
Let ˆp
(h)
nkr
denote the OOB predicted probability that a heuristic
h assigns to an observation n of belonging to a response class k in an experimental replication
r. Then, within each experimental replication r, we can compare the predicted probabilities
that two different heuristics h
1
, h
2
∈ H assign to an observation n by considering the
difference ˆp
(h
1
)
nkr
− ˆp
(h
2
)
nkr
. We summarize these differences in the predicted probabilities of
voting “Yes” for all possible pairwise combinations of the seven heuristics in Figure 4, where
each panel plots the mean and middle 95% of these differences across all 1000 experimental
replications as a function of the OOB absence proportion.
The large discrepancies in the predicted probabilities that are observed in Figure 4 are
particularly concerning since they can lead to different classifications. If we let ˆy
(h)
nr
denote
the OOB classification that a heuristic h makes for an observation n in an experimental
replication r, then Cohen’s kappa coefficient (Cohen, 1960) provides one way of measuring
the level of agreement between two different heuristics h
1
, h
2
∈ H:
κ
(h
1
,h
2
)
r
=
o
(h
1
,h
2
)
r
− e
(h
1
,h
2
)
r
1 − e
(h
1
,h
2
)
r
, (14)
where
o
(h
1
,h
2
)
r
=
1
N
N
X
n=1
I
ˆy
(h
1
)
nr
= ˆy
(h
2
)
nr
is the observed probability of agreement between the two heuristics, and where
e
(h
1
,h
2
)
r
=
1
N
2
K
X
k=1
"
N
X
n=1
I
ˆy
(h
1
)
nr
= k
!
·
N
X
n=1
I
ˆy
(h
2
)
nr
= k
!#
is the expected probability of the two heuristics agreeing by chance. Therefore, within an
experimental replication r, we will observe κ
(h
1
,h
2
)
r
= 1 if the two heuristics are in com-
plete agreement, and we will observe κ
(h
1
,h
2
)
r
≈ 0 if there is no agreement amongst the two
heuristics other than what would be expected by chance. In Figure 5, we plot histograms
of the Cohen’s kappa coefficient for all possible pairwise combinations of the seven heuris-
tics across all 1000 experimental replications when the random forests algorithm’s default
majority vote discrimination threshold of 0.5 is used.
More generally, the areas underneath the receiver operating characteristic (ROC) and
precision-recall (PR) curves can be used to compare the overall performance of binary
classifiers as the discrimination threshold is varied between 0 and 1. Specifically, as the dis-
crimination threshold changes, the ROC curve plots the proportion of positive observations
that a classifier correctly labels as a function of the proportion of negative observations
that a classifier incorrectly labels, while the PR curve plots the proportion of a classifier’s
positive labels that are truly positive as a function of the proportion of positive observations
that a classifier correctly labels (Davis and Goadrich, 2006).
9
This is the approach that is used by the randomForest R package. The scikit-learn Python module
uses an alternative method of calculating the predicted response class probabilities which takes the average
of the predicted class probabilities over the trees in the random forest, where the predicted probability of
a response class k in an individual tree is estimated using the proportion of a node’s training samples that
belong to the response class k.
22
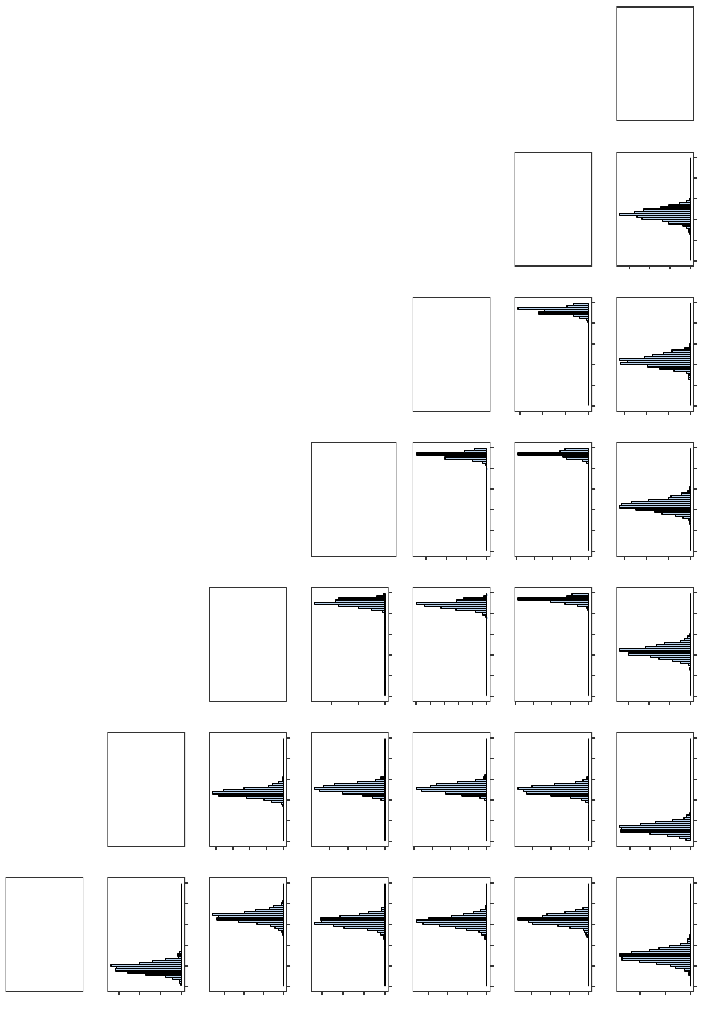
The “Absent Levels” Problem
Left
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
Right
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
0
50
100
150
200
0.5 0.6 0.7 0.8 0.9 1.0
Stop
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
0
100
200
0.5 0.6 0.7 0.8 0.9 1.0
Majority
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
0
50
100
150
200
0.5 0.6 0.7 0.8 0.9 1.0
0
50
100
150
200
250
0.5 0.6 0.7 0.8 0.9 1.0
0
100
200
300
0.5 0.6 0.7 0.8 0.9 1.0
Random
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
0
100
200
300
400
0.5 0.6 0.7 0.8 0.9 1.0
0
100
200
300
400
0.5 0.6 0.7 0.8 0.9 1.0
0
100
200
300
0.5 0.6 0.7 0.8 0.9 1.0
DBI
0
50
100
0.5 0.6 0.7 0.8 0.9 1.0
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
0
50
100
150
0.5 0.6 0.7 0.8 0.9 1.0
One−Hot
Cohen's Kappa
Frequency
Figure 5: Pairwise Cohen’s kappa coefficients for the seven different heuristics as defined in (14) when the random forests algo-
rithm’s default majority vote discrimination threshold of 0.5 is used in the PROMESA data set. Each panel plots the
histogram of coefficients across all 1000 experimental replications when the OOB classifications of the heuristic that
is labeled at the right of the panel’s row are compared against the OOB classifications of the heuristic that is labeled
at the top of the panel’s column. Cohen’s kappa coefficients were calculated within each of the 1000 experimental
replications to account for the positive correlation between the naive and missing data heuristics.
23

Au
Taking the “Yes” vote to be the positive response class in our analysis, we calculate the
areas underneath the ROC and PR curves for each heuristic h ∈ H within each experimental
replication r. Boxplots depicting each heuristic’s marginal distribution of these two areas
across all 1000 experimental replications are shown in the left panels of Figure 6. However,
these marginal boxplots ignore the positive correlation that exists between the naive and
missing data heuristics. Therefore, within every experimental replication r and similar to
what was previously done in our 1985 Auto Imports example, we also compare the areas
for each heuristic h ∈ H relative to the best area that was achieved amongst the missing
data heuristics H
m
= {Stop, Majority, Random, DBI }:
AUC
(hkH
m
)
r
=
AUC
(h)
r
− max
h∈H
m
AUC
(h)
r
max
h∈H
m
AUC
(h)
r
, (15)
where, depending on the context, AUC
(h)
r
denotes the area that is underneath either the
ROC or PR curve for heuristic h in experimental replication r. Boxplots of these relative
areas across all 1000 experimental replications are displayed in the right panels of Figure 6.
5.2.1 Naive Heuristics
As expected given our discussions in Section 3.2 and how we have chosen to index the
response classes in our analysis, we see from Figure 4 that the Left heuristic results in sig-
nificantly higher predicted probabilities of voting “Yes” than the other heuristics, while the
Right heuristic yields predicted probabilities of voting “Yes” that are substantially lower.
The consequences of this behavior in terms of making classifications can be observed in
Figure 5, where we note that both the Left and Right heuristics tend to exhibit a high level
of disagreement when compared against any other heuristic’s classifications. Moreover, Fig-
ure 6 illustrates that the randomForest R package’s practice of always sending absent levels
left in binary classification is noticeably detrimental here—relative to the best performing
missing data heuristic, it gives areas underneath the ROC and PR curves that are, on av-
erage, 1.5% and 3.7% worse, respectively. And although the Right heuristic appears to do
well in terms of the area underneath the PR curve, we once again emphasize the spurious
nature of this performance and caution against taking it at face value.
5.2.2 Missing Data Heuristics
Similar to what was previously seen in our 1985 Auto Imports example, Figures 4 and 5
show that the four missing data heuristics tend to exhibit a higher level of agreement with
one another than they do with the Left, Right, and One-Hot heuristics. However, significant
differences do still exist, and we see from Figure 6 that the relative performances of the
heuristics will vary depending on the specific task at hand—the Majority heuristic slightly
outperforms the three other missing data heuristics in terms of the area underneath the
ROC curve, while the Random heuristic does considerably better than all of its missing
data counterparts with respect to the area underneath the PR curve.
24
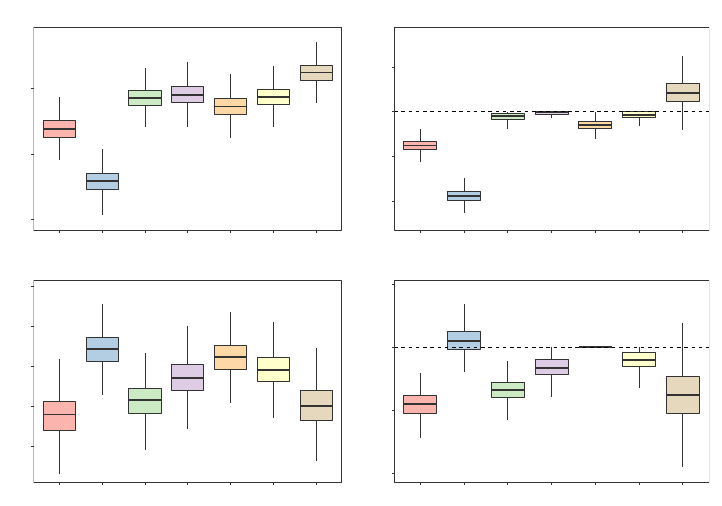
The “Absent Levels” Problem
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
0.68
0.70
0.72
Left Right Stop Majority Random DBI One−Hot
Receiver Operating Characteristic (ROC) Curve
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
0.76
0.78
0.80
0.82
0.84
Left Right Stop Majority Random DBI One−Hot
Precision−Recall (PR) Curve
Area
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
−4
−2
0
2
Left Right Stop Majority Random DBI One−Hot
Receiver Operating Characteristic (ROC) Curve
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
−8
−4
0
4
Left Right Stop Majority Random DBI One−Hot
Precision−Recall (PR) Curve
Relative Area (%)
Absent Levels Heuristic
Figure 6: Areas underneath the ROC and PR curves for the seven heuristics in the
PROMESA data set. The left panels show boxplots of each heuristic’s marginal
distribution of areas across all 1000 experimental replications, which ignores the
positive correlation that exists between the naive and missing data heuristics.
The right panels account for this positive correlation by comparing the areas of
the heuristics relative to the best area that was obtained amongst the missing
data heuristics within each of the 1000 experimental replications as in (15).
5.2.3 Feature Engineering Heuristic
Although it is essentially uncorrelated with the other six heuristics across all 1000 exper-
imental replications, Figures 4 and 5 still suggest that the One-Hot heuristic’s predicted
probabilities and classifications can greatly differ from the other six heuristics. Further-
more, Figure 6 shows that even though the One-Hot heuristic may appear to perform well
in terms of the area underneath its ROC curve relative to the missing data heuristics, its
performance in the PR context is rather lackluster.
5.3 Pittsburgh Bridges
For a multiclass classification example, we consider the Pittsburgh Bridges data set from
the UCI Machine Learning Repository which, after removing observations with missing
data, contains seven predictors that can be used to classify 72 bridges to one of seven
different bridge types. The categorical predictors in this data set for which the absent levels
problem can occur include a bridge’s river (3 levels), purpose (3 levels), and location (46
25

Au
levels). Consequently, recall from Section 3.2, that the random forests FORTRAN code and
randomForest R package will both employ an exhaustive search that always sends absent
levels right when splitting on either the river or purpose predictors, and that they will
both resort to using a random search that sends absent levels either left or right with equal
probability when splitting on the location predictor since it has too many levels for an
exhaustive search to be computationally efficient. The OOB absence proportions for this
example are summarized in the bottom panel of Figure 1 and in Table 1.
Within each experimental replication r, we can use the log loss
LogLoss
(h)
r
= −
1
N
N
X
n=1
K
X
k=1
h
I(y
n
= k) · log
ˆp
(h)
nkr
i
to evaluate the overall performance of each heuristic h ∈ H, where we once again let
ˆp
(h)
nkr
denote the OOB predicted probability that a heuristic h assigns to an observation
n of belonging to a response class k in an experimental replication r. The left panel of
Figure 7 displays the marginal distribution of each heuristic’s log losses across all 1000
experimental replications. However, to once again account for the positive correlation that
exists amongst the naive and missing data heuristics, within every experimental replication
r, we also compare the log losses for each heuristic h ∈ H relative to the best log loss that
was achieved amongst the missing data heuristics H
m
= {Stop, Majority, Random, DBI }:
LogLoss
(hkH
m
)
r
=
LogLoss
(h)
r
− min
h∈H
m
LogLoss
(h)
r
min
h∈H
m
LogLoss
(h)
r
. (16)
Boxplots of these relative log losses are depicted in the right panel of Figure 7.
5.3.1 Naive Heuristics
Although we once again stress the systematically biased nature of the Left and Right heuris-
tics, we note from Figure 7 that the two naive heuristics are sometimes able to outperform
the missing data heuristics. Nevertheless, on average, the Left and Right heuristics resulted
in log losses that are 0.7% and 1.9% worse than the best performing missing data heuristic,
respectively.
5.3.2 Missing Data Heuristics
Figure 7 shows that for this particular example, the Majority and Random heuristics per-
form roughly on par with one another, and that they both also significantly outperform
the Stop and DBI heuristics—the smallest log loss amongst all of the missing data heuris-
tics was achieved by either the Majority or the Random heuristic in 999 out of the 1000
experimental replications.
5.3.3 Feature Engineering Heuristic
It can also be observed from Figure 7 that, although the One-Hot heuristic can occasionally
outperform the missing data heuristics, on average, it yields a log loss that is 4.5% worse
than the best performing missing data heuristic.
26
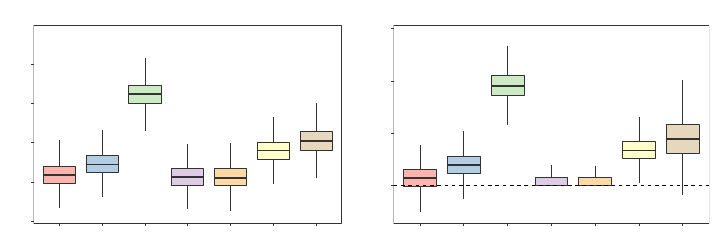
The “Absent Levels” Problem
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
1.10
1.15
1.20
1.25
1.30
Left Right Stop Majority Random DBI OneHot
Log Loss
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
0
5
10
15
Left Right Stop Majority Random DBI OneHot
Relative Log Loss (%)
Absent Levels Heuristic
Figure 7: Log losses for the OOB predicted response class probabilities of the seven heuris-
tics in the Pittsburgh Bridges data. The left panel shows boxplots of each heuris-
tic’s marginal distribution of log losses set across all 1000 experimental replica-
tions, which ignores the positive correlation that exists between the naive and
missing data heuristics. The right panel accounts for this positive correlation by
comparing the log losses of the heuristics relative to the best log loss that was ob-
tained amongst the missing data heuristics within each of the 1000 experimental
replications as in (16).
6. Conclusion
In this paper, we introduced and investigated the absent levels problem for decision tree
based methods. In particular, by using Breiman and Cutler’s random forests FORTRAN code
and the randomForest R package as motivating case studies, we showed how overlooking the
absent levels problem could systematically bias a model. Furthermore, we presented three
real data examples which illustrated how absent levels can dramatically alter a model’s
performance in practice.
Even though a comprehensive theoretical analysis of the absent levels problem was
beyond the scope of this paper, we empirically demonstrated how some simple heuristics
could be used to help mitigate the effects of absent levels. And although none of the missing
data and feature engineering heuristics that we considered performed uniformly better than
all of the others, they were all shown to be superior to the biased naive approaches that
are currently being employed due to oversights in the software implementations of decision
tree based methods.
Consequently, until a more robust theoretical solution is found, we encourage the soft-
ware implementations which support the native categorical split capabilities of decision trees
to incorporate the Random heuristic as a provisional measure given its reliability—in all of
our examples, the Random heuristic was always competitive in terms of its performance.
Moreover, based on our own personal experiences, we note that the Random heuristic was
one of the easier heuristics to implement on top of the randomForest R package. In the
meantime, while waiting for these mitigations to materialize, we also urge users who rely
on decision tree based methods to feature engineer their data sets when possible in order to
circumvent the absent levels problem—although our empirical results suggest that this may
27
Au
sometimes be detrimental to a model’s performance, we believe this to still be preferable
to the alternative of having to rely on biased approaches which do not adequately address
absent levels.
Finally, although this paper primarily focused on the absent levels problem for random
forests and a particular subset of the types of analyses in which random forests have been
used, it is important to recognize that the issue of absent levels applies much more broadly.
For example, decision tree based methods have also been employed for clustering, detecting
outliers, imputing missing values, and generating variable importance measures (Breiman,
2003)—tasks which also depend on the terminal node behavior of the observations. In ad-
dition, several extensions of decision tree based methods have been built on top of software
which currently overlook absent levels—such as the quantile regression forests algorithm
(Meinshausen, 2006, 2012) and the infinitesimal jackknife method for estimating the vari-
ance of bagged predictors (Wager et al., 2014), which are both implemented on top of the
randomForest R package. Indeed, given how extensively decision tree based methods have
been used, a sizable number of these models have almost surely been significantly and un-
knowingly affected by the absent levels problem in practice—further emphasizing the need
for the development of both theory and software that accounts for this issue.
Acknowledgements
The author is extremely grateful to Art Owen for numerous valuable discussions and in-
sightful comments which substantially improved this paper. The author would also like to
thank David Chan, Robert Bell, the action editor, and the anonymous reviewers for their
helpful feedback. Finally, the author would like to thank Jim Koehler, Tim Hesterberg,
Joseph Kelly, Iv´an D´ıaz, Jingang Miao, and Aiyou Chen for many interesting discussions.
References
Yali Amit and Donald Geman. Shape quantization and recognition with randomized trees.
Neural Computation, 9:1545–1588, 1997.
G´erard Biau. Analysis of a random forests model. Journal of Machine Learning Research,
13(1):1063–1095, 2012.
G´erard Biau, Luc Devroye, and G´abor Lugosi. Consistency of random forests and other
averaging classifiers. Journal of Machine Learning Research, 9(1):2015–2033, 2008.
Leo Breiman. Bagging predictors. Machine Learning, 24(2):123–140, 1996a.
Leo Breiman. Out-of-bag estimation. Technical report, Department of Statistics, U.C.
Berkeley, 1996b. URL https://www.stat.berkeley.edu/
~
breiman/OOBestimation.
pdf.
Leo Breiman. Random forests. Machine Learning, 45(1):5–32, 2001.
Leo Breiman. Manual—setting up, using, and understanding random forest v4.0. 2003.
URL https://www.stat.berkeley.edu/
~
breiman/Using_random_forests_v4.0.pdf.
28
The “Absent Levels” Problem
Leo Breiman, Jerome. H. Friedman, Richard. A. Olshen, and Charles J. Stone. Classification
and Regression Trees. Wadsworth, 1984. ISBN 0-534-98053-8.
Jacob Cohen. A coefficient of agreement for nominal scales. Educational and Psychological
Measurement, 20:37–46, 1960.
Jesse Davis and Mark Goadrich. The relationship between precision-recall and ROC curves.
In ICML ’06: Proceedings of the 23rd international conference on Machine learning, pages
233–240, 2006.
Misha Denil, David Matheson, and Nando de Freitas. Narrowing the gap: Random forests
in theory and in practice. In Proceedings of the 31th International Conference on Machine
Learning, pages 665–673, 2014.
Walter D. Fisher. On grouping for maximum homogeneity. Journal of the American Sta-
tistical Association, 53(284):789–798, 1958.
Trevor J Hastie, Robert J. Tibshirani, and Jerome H. Friedman. The Elements of Statistical
Learning: Data Mining, Inference, and Prediction, Second Edition. Springer Series in
Statistics. Springer, New York, 2009.
Tin Kam Ho. The random subspace method for constructing decision forests. IEEE Trans-
actions on Pattern Analysis and Machine Intelligence, 20(8):832–844, 1998.
Torsten Hothorn and Achim Zeileis. partykit: A modular toolkit for recursive partytioning
in R. Journal of Machine Learning Research, 16:3905–3909, 2015.
Torsten Hothorn, Kurt Hornik, and Achim Zeileis. Unbiased recursive partitioning: A
conditional inference framework. Journal of Computational and Graphical Statistics, 15
(3):651–674, 2006.
Hemant Ishwaran, Udaya B. Kogalur, Eugene H. Blackstone, and Michael S. Lauer. Random
survival forests. Annals of Applied Statistics, 2(3):841–860, 2008.
Jeff Lewis. Rvoteview: Voteview Data in R, 2015. https://github.com/JeffreyBLewis/
Rvoteview, http://voteview.polisci.ucla.edu, http://voteview.com.
Andy Liaw and Matthew Wiener. Classification and regression by randomForest. R News,
2(3):18–22, 2002.
Moshe Lichman. UCI machine learning repository, 2013. URL http://archive.ics.uci.
edu/ml.
Wei-Yin Loh and Nunta Vanichsetakul. Tree-structured classification via generalized dis-
criminant analysis. Journal of the American Statistical Association, 83(403):715–725,
1988.
Nolan M. McCarty, Keith T. Poole, and Howard Rosenthal. Income Redistribution and
the Realignment of American Politics. AEI Press, publisher for the American Enterprise
Institute, 1997.
29
Au
Nicolai Meinshausen. Quantile regression forests. Journal of Machine Learning Research,
7:983–999, 2006.
Nicolai Meinshausen. quantregForest: Quantile Regression Forests, 2012. URL http://
CRAN.R-project.org/package=quantregForest. R package version 0.2-3.
Fabian Pedregosa, Ga¨el Varoquaux, Alexandre Gramfort, Vincent Michel, Bertrand
Thirion, Olivier Grisel, Mathieu Blondel, Peter Prettenhofer, Ron Weiss, Vincent
Dubourg, Jake Vanderplas, Alexandre Passos, David Cournapeau, Matthieu Brucher,
Matthieu Perrot, and
´
Edouard Duchesnay. Scikit-learn: Machine learning in Python.
Journal of Machine Learning Research, 12:2825–2830, 2011.
John R. Quinlan. C4.5: Programs for Machine Learning. Morgan Kaufmann Publishers
Inc., 1993.
Greg Ridgeway. gbm: Generalized Boosted Regression Models, 2013. URL http://CRAN.
R-project.org/package=gbm. R package version 2.1.
Brian D. Ripley. Pattern Recognition and Neural Networks. Cambridge University Press,
Cambridge, 1996. ISBN 0-521-46086-7.
Maytal Saar-Tsechansky and Foster Provost. Handling missing values when applying clas-
sification models. Journal of Machine Learning Research, 8:1623–1657, 2007.
Terry Therneau, Beth Atkinson, and Brian Ripley. rpart: Recursive Partitioning and Re-
gression Trees, 2015. URL http://CRAN.R-project.org/package=rpart. R package
version 4.1-10.
Stefan Wager, Trevor Hastie, and Bradley Efron. Confidence intervals for random forests:
The jackknife and the infinitesimal jackknife. Journal of Machine Learning Research, 15
(1):1625–1651, 2014.
30
