
Jean-Christophe.Mozziconacci.and.Volker.Eyrich!
http://www.schrodinger.com/upload/KNIME_Overview.pdf!!
KNIME desktop and Schrödinger extensions overview

Schrödinger extensions
• 150+ nodes:
– Covering most of the Schrödinger tools
Most recent additions: Residue scanning, Prime Energy, SiteMap, PyMOL, Glide grid writer
Many take in and output sdf and pdb or mol2 on top of Maestro format
Newest nodes use the same configuration panel as Maestro (see the Residue scanning node)
– Structure and data manipulation nodes (eg Split by structure, delete atoms)
– GUI nodes (eg Run Maestro, Run PyMOL)
– Scripting nodes: Run Maestro command, Chemistry external tool, Python nodes
– Utility nodes: Setup diagnosis and workflow list
• 50+ workflow examples
– Workflow page: descriptions and download the ones of interest
– Whole set can be downloaded with the Suite
– Many other workflow drafts available on demand

Schrödinger extensions
• Tested with latest KNIME version and include the version available at the time
of the release
• Parameter flow variable capability
– Use the options not exposed in the configuration panels (eg command line only)
– Implemented for the main nodes eg Glide, MMod (eg OPLS 2.1), Jaguar
• 2D renderer
– 2D coordinate generation and rendering
The default renderer can be set for Maestro columns
– Can be used in the Report designer

Schrödinger extensions
• More and simplified start-up options and stand-alone installation configuration in the Preferences
– In $SCHRODINGER/knime start-up script
– eg use a stand-alone KNIME installation, set temporary directories, the memory limit…
• KNIME menu in Maestro
– Connect to KNIME mode to exchange structures with a KNIME session
KNIME-Maestro connector node (Improved in 2015-1: automatic connection/close, more modes)
– Build, import, edit and run workflows from Maestro on project table data
Dynamically generated GUI to alter some parameters.
• Simplified batch execution: KNIME_batch.py
– Batch command generation based on workflow annotations
Stand-alone dynamically generated GUI
– Useful options eg -stderr/out
• Easier installation creation and update
– Using Eclipse machinery in KNIME_install.py
– eg list of extensions to install, from several (zipped) update sites

Schrödinger extensions
See details in:
http://www.schrodinger.com/upload/KNIME_Overview.pdf
• The new features slides:
– Parameter flow variables (2014-1)
– Simplified batch execution (2013, 2014-1)
– Chemistry external tool node improvements
– KNIME menu in Maestro (2012)
– Start-up script (2012)
• Workflow examples:
– Labs > Parameter flow variable usage
– General > Installation (KNIME_install.py scenarios)
– General > Workflow list
– General > Chemistry external tool node usage
– Phase > Shape screening, docking (batch execution)
– 2D renderer in the report designer

Why use KNIME
Automation
- Automate repetitive tasks (especially complex / error prone
tasks)
- e.g. PDB set preparation, Ligand database preparation
Collaboration
- Share workflows with colleagues (also in Maestro and Seurat)
- Not a black box
- e.g. Ensemble docking, HTS analysis
Prototyping
- Test parameters
- e.g. Validate docking parameters
Documentation
- Easy debugging, interruptions, data included, inspect each step
(2D, in Maestro/PyMOL)
Combine various Tools
- Schrodinger and third party tools (vendor agnostic), scripts
(Python, Java, shell),
- e.g. QM workflows
Reporting
- Nodes for reporting results
- e.g. Enrichment plotter node, pdf, tables, etc.
- KNIME report designer (free), web portal (KNIME.com)
Inexpensive

Overview
• Organized by level:
– Get started
– Intermediate
– Advanced functionalities
• And by topics:
– KNIME desktop: GUI, specificities, nodes
– Schrödinger extensions: specificities, nodes
• You can jump between the sections using links (marked with ► or◄). See the
overview slides.
• There are also links to use-case examples (marked with ♦).

Get started
KNIME desktop
– GUI ►
– Specificities ►
– Nodes ►
Schrödinger extensions
– Specificities ►
– Schrödinger nodes ►
►!Intermediate!
!

KNIME desktop GUI ◄
• Knime.org and Knime.com
• KNIME desktop
• Start Knime
• Create a new workflow and organize a workspace
• Run a node
• Import and export workflows
• Tips and tricks
• Documentation

Konstanz Information Miner and Ecosystem
KNIME.org
- Leading open-source ‘pipelining/workflow’ tool
- Freely available to academic and industrial researchers
- KNIME Desktop, based on Eclipse !http://www.knime.org!
- Community contributions:
- Modeling tools
- Marvin sketcher
- RDKit
- Indigo
- CDK
- R Scripting
- Erlwood
- Image Processing
- HCS Tools
- Next Generation Sequencing
- Palladian (mainly GPL3)
KNIME.com Enterprise products and Services !http://www.knime.com!
- Report designer (free)
- KNIME team space (share workflows) and KNIME server (web portal and SOA access)
- Cluster execution (scalability)
- KNIME professional (support, maintenance and training)
- Commercial development

KNIME Extensions
15+ Extension Providers
Extensible, cross-platform, vendor neutral:
Schrödinger, CCG, Tripos, ChemAxon/Infocom, BioSolveIT, Cresset, Dotmatics, Molecular
Discovery, Molegro…
Schrödinger Extensions
- First released in 2007
- 150+ nodes
- Molecular mechanics
- Molecular dynamics
- Quantum mechanics
- Cheminformatics
- Pharmacophore modeling
- Combinatorial libraries
- Docking
- Protein structure prediction
- Structure and data manipulation
- Maestro integration
- Workflow execution
- Structure exchange

KNIME Desktop GUI
• Full screen mode
• Forget about Eclipse
specific menu items
More about:
• The console ►

Start KNIME
• Start up KNIME:
– On Linux: run $SCHRODINGER/knime
– On Windows: click on the icon
– Use -data MyWorkspace to open a specific workspace
– File > Switch workspace, but KNIME takes time to start up again
• Workspace, workflows and workflow groups:

Create a new workflow and organize a workspace
• Under the pop-up menu of Workflow Project repository:
– New KNIME workflow and New Workflow group
– Copy, Paste, Delete, Move, Rename
• Drag and drop the workflows in the Workflow project repository

Run a node
double!click!or!
pop-up!
• Connectors
• Node status
• Input!and!output!
data!tables!

Import and export workflows
• File > Import KNIME workflow / Export KNIME workflow
or under the pop-up menu of a Workflow Project group
• Import from another workspace or an archive file (zip)
• Select 1 or several workflows > export as a zip file
Exclude or not the cached data from the exported file

Tips and tricks
• Save regularly the changes. Since KNIME 2.10 there is an auto save functionality but it isn’t on
by default.
Save while running calculations (see Preferences ►)
• Multiple undo and redo apply on workflow edition (execution data lost though)
• The KNIME desktop isn’t based on a client-server architecture. If you close the KNIME instance
while a calculation is running it won’t kill it nor you will be able to recover the results when
opening the workflow again.

Documentation
• Node descriptions. Also accessible from Help > Help content > Knime > Node descriptions
Type in search field and inspect the configuration panel
• Product page http://www.schrodinger.com/KNIME-Extensions
New Features, New Features Slides, KNIME Overview
• Schrödinger KNIME manual $SCHRODINGER/docs/knime/user_manual/kni13_user_manual.pdf
• Schrödinger FAQs http://www.schrodinger.com/kb
• Workflow page (examples) http://www.schrodinger.com/knimeworkflows/
• Schrödinger’s extensions webex http://www.schrodinger.com/seminarprior/19/24/
• KNIME.org
– Workflow examples (preconfigured server access in KNIME explorer)
– Screencast http://www.knime.com/introduction/screencasts
– Forum http://www.knime.org/forum/
KNIME desktop GUI ►
• Preferences
• Advanced node functionalities
• Errors, warnings and Console information
• Flow variables and workflow variables
• Metanodes
• Memory limit
• Tips and tricks

KNIME desktop specificities ◄
• Stepwise execution
• Data table column types and conversion

Stepwise execution
• Only stepwise execution
Ideal to take advantage of Schrödinger’s jobcontrol infrastructure
• No predefined execution order for non connected branches. Use the flow variable ports
[KNIME 2.3]
• Data cached at each step
• Preferences > KNIME > Maximum working threads far all nodes
• 1:1 connection between nodes (use the concatenate node to combine input flows)

Data table column types and conversion
• KNIME relies on strict data table column typing
• Converter nodes:
– Double to Int (integer), String to number, Number to string
– Molecule type cast (but no Maestro conversion)
– Openbabel, CDK to molecule, Molecule to CDK
• In the Schrödinger extensions:
– String-to-type
– Molecule-to-MAE, MAE-to-Pdb, MAE-to-SD, MAE-to-Smiles, MAE-to-mol2, SD-to-smiles
– Canvas object converters

Schrödinger specific cell types
• Structures: Maestro, Sequence, Alignment
• Several files: Glide grid, Phase Hypothesis
• Desmond trajectory
• Binary formats: Canvas fingerprint and matrix
Readers and writers, converters

KNIME workbench nodes ◄
• KNIME workbench nodes
• Data manipulation nodes
• Data exchange

KNIME workbench nodes
• I/O nodes for reading and writing data from files and databases
• Data manipulation nodes for managing the internal data tables that are
used to pass information between nodes
• Charting and plotting tools
• Loop support, time Series, Distance matrix
• Statistics and data mining nodes (Mining, Weka) such as clustering, neural
networks, decision trees, Lib SVM
R statistical computation
• Basic chemistry-aware nodes (CDK)
very limited, see Schrödinger extension nodes ►

The most often used nodes for data manipulation
• Row filter, Row splitter and Sorter
• Column filter, Column resorter, Column combiner and Rename
• Joiner (see also Schrödinger Look up and add column node ►) and Concatenate (only 2 inputs)
And also:
• Java snippet, RowID and GroupBy node ►
• Schrödinger nodes for data manipulation ►

Data exchange
• As text files: File reader and csv writer nodes
• In Excel format: xls reader and xls writer nodes
• Between workflows: table reader and table writer nodes
• See also among the Schrödinger nodes:
– Schrödinger reader and writer nodes
– CSV reader (read several files)
– View CSV ►

KNIME workbench nodes ►
• KNIME.com Labs nodes
• Scripting and run a third party tool
• Java snippet
• RowID
• Group by
• Miscellaneous nodes: Interactive table, Math formula, CDK Sketcher
• Plotting facilities
• Looping functionalities - Basics
• Model building nodes

Schrödinger extensions specificities ◄
• Canvas 2D renderer
• Grouped structures in a cell
• Output column structure options
• Jobcontrol tab

Canvas 2D renderer
• Preferences > KNIME > Preferred renderer

Grouped structures in a cell
• #CTs: number of structures
• Set of conformations, Glide poses, Ligprep forms…
• Group and ungroup nodes, match option
• Also grouped SD, mol2

Output column structure options
• Input plus Output, Output replaces Input, Output only
• Extract MAE properties, Set MAE properties and delete
MAE properties nodes
CTs:
Structures + properties
CTs, columns new CTs and/or new columns
KNIME
Schrödinger
tools
New CTs
Extract mae
properties
Set mae properties
Reader
Writer,
Maestro
Delete mae
properties

Output column structure options

Jobcontrol

Schrödinger extensions specificities ►
• Schrödinger preferences
• Start-up script options
• Access to flow variables

Schrödinger nodes ◄
• Schrödinger node repository
• Configuration panel visual coherence
• Nodes of general use:
– Readers and converters
– Run Maestro and Run Maestro command
– Structure manipulation
– Data manipulation and viewers
– Scripting
• KNIME workflow webpage

Access to Schrödinger tools via KNIME
• Run on Linux, Mac and Windows
32 and 64 bit
• Add our extensions to an existing KNIME
installation using the update site
• 150+ nodes covering the whole
Schrödinger Suite
Cheminformatics.
.Fingerprint.Based.Tools.
-!Fingerprint!Generation!
-!Generate!Pairwise!Matrix!
-!Generate!Pairwise!Matrix!(2!Inputs)!
-!Similarity!Matrix!(from!Molecules)!
-!Dissimilarity!Selection!(from!Matrix)!
-!Build!Report!for!Clustering!(from!Matrix)!
-!Hierarchical!Clustering!(from!Matrix)!
.Filters.and.Mining.Tools.
-!Maximum!Common!Substructure!Search!
-!Substructure!Search!
-!REOS!Filter!
-!Structure!Filter!
.Utilities.and.Converters.
-!Principal!Components!
-!Multi-dimensional!Scaling!
-!Combine!Fingerprints!
-!Concatenate!Bitvectors!
-!Convert!Fingerprint!to!Bitvector!
-!Convert!Fingerprint!to!Table!
-!Convert!Matrix!to!Table!
-!Convert!Table!to!FingerPrint!
-!Convert!Table!to!Matrix!
-!Convert!Bitvector!to!FingerPrint!
.Modeling.
-!Bayes!Classification!Model!Building!
!
!
!
!
Molecular.Mechanics.
-!MacroModel!Single!Point!Energy!
-!MacroModel!Minimization!
-!MacroModel!Coordinate!Scan!
-!ConfGen!Standard!
-!ConfGen!
-!Conformational!Search!
-!Conformational!Search!and!Cluster!
-!Premin!
-!Impref!
-!Uffmin!
Quantum.Mechanics.
-!Jaguar!Single!Point!Energy!
-!Jaguar!Minimization!
-!NMR!Shielding!Constants!
-!Jaguar!Charges!
!
Pharmacophore.Modeling.
-!Phase!Shape!
-!Phase!DB!Query!
-!Phase!File!Query!
-!Phase!DB!Creation!
-!Phase!Hypothesis!Identification!
Docking.and.Scoring.
-!Glide!Grid!Generation!
-!Glide!Ligand!Docking!
-!Glide!Multiple!Ligand!Docking!
-!XP!Visualizer!
.Post-processing.
-!Prime!MM-GBSA!
-!Embrace!Minimization!
-!Strain!Rescore!
-!Pose!Entropy!
-!Pose!Filter!
-!Glide!Ensemble!Merge!
-!Glide!Merge!
-!Glide!Sort!Results!
.
!
!
!
!
!
!
!
Protein.Structure.Prediction.
-!BLAST!
-!Prime!Build!Homology!Model!
-!Prime!Side!Chain!Sampling!
-!Prime!Minimization!

Schrödinger nodes
Workflows.
!!!!!Protein!Preparation!
-!Protein!Preparation!Wizard!
-!Protein!Assignment!
!!!!!Induced-fit!docking!
-!IFD!and!individual!steps!
Ligand.Preparation.
-!LigPrep!
-!Ligprep!individual!tools!(Ionizer,!
Desalter,!Neutralizer…)!
-!Epik!
Property.Generation.
-!QikProp!
-!Molecular!Descriptors!
-!Calculate!properties!
Filtering.
-!Ligfilter!
-!Ligparse!
-!Property!Filter!(Propfilter).
Readers/Writers.
-!CSV!Reader!
-!Molecule!Reader!
-!SD,!PDB,!Mol2!Reader!nodes!
-!Sequence!Reader!
-!Alignment!Reader!
-!Fingerprint!Reader!
-!Hypothesis!Reader!
-!Glide!Grid!Reader!
-!Glide!Multiple!Grid!Reader!
-!Variable!Based!Glide!Grid!Reader!
-!Molecule!Writer!
-!Sequence!Writer!
-!Alignment!Writer!
-!Hypothesis!Writer!
-!Fingerprint!Writer!
Converters.
-!Molecule-to-MAE!
-!MAE-to-Pdb,!to-SD,!to-Smiles!and!to-Mol2!
-!SD-to-Smiles!
-!PoseViewer-to-Complexes!
-!Complexes-to-PoseViewers!
-!String-to-Type!
-!Hartree-to-kcal/mol!Converter!
-!kJ-to-kcal!Converter!
!
!
!
!
!
!
Desmond.
-!System!builder!
-!Molecular!Dynamics!
-!Trajectory!extract!frames!and!manipulation!
-!Trajectory!reader,!CMS!reader!
Reporting.
-!Run!Maestro!
-!Run!Canvas!
-!View!CSV!(open!xls/ooffice)!
-!Text!Viewer!
!
Tools.
.....Combinatorial.Libraries.
-!CombiGlide!Library!Enumeration!
-!CombiGlide!Reagent!Preparation!
.....Fragments.
-!Fragment!Joiner!
-!Fragments!from!Molecules!
!!!!!Data.Manipulation.
-!Compare.Ligands.
-!Lookup!and!Add!Columns!
-!Group.MAE.
-!Ungroup!MAE!
!
.
!!
.....!
Structure.Manipulation.
-!Add!Hydrogens!
-!Delete.Atoms.
-!Split.by.Structure.
-!MAE.Parser.
-!Extract!MAE!Properties!
-!Delete!MAE!Properties!
-!Set!MAE!Properties!
-!Set!Molecule!Title!
-!Set!MAE!Index!
!!!!Utilities.
-!Get!PDB!
-!Align!Binding!Sites!
-!Protein!Structure!Alignment!
-!Prime!Fix!
-!RMSD.
-!Assign!Bond!Orders!
-!Unique!Title!Check!
-!PDB!Name!
-!SD!Format!Checker!
-!Generate!Smarts!
-!Unique!Smiles!
-!Entropy!Calculation!
-!RRHO!Entropy!
-!Boltzmann!Population!
-!Volume!Overlap!Matrix!
Scripting.
-!Run.Maestro.Command.
-!Chemistry.External.Tool.0:1,!1:0,!1:1,!
1:2,!2:1!and!2:2!nodes!
-!Python.Script.0:1,!1:0,!1:1,!!1:2,!2:1!and!
2:2!nodes!
!
!
!
!
!
!

Visual Coherence – Maestro vs. KNIME
Read in
Ligands
Ligand
Preparation
Filtering
Read in
Grid
Docking
View
Results

Nodes of general use - Readers and converters
• Molecule reader, SD reader... Glide grid reader...
• Converters (Maestro, mae.gz, SD, sd.gz, mol2, PDB, smiles) including Molecule to MAE,
string to type. Canvas converters (Matrix, Fingerprint, Bitvector from and to table). SD
format checker
• Pose viewer to complexes and Complexes to PoseViewer

Run maestro command and Run Maestro

Nodes of general use - Structure manipulation
• Set MAE properties
• Extract!MAE!properties!

Nodes of general use - Structure manipulation
• Extract and Set MAE properties
• Group and Ungroup, Set MAE index
• MAE parser
• Split by structure, Delete atoms
• Compare ligands
• Unique smiles, Unique title check, Ligfilter,
Align binding sites, RMSD,
Volume overlap matrix...

Nodes of general use - Data manipulation and viewers
• Look up and add columns
• Run Spreadsheet viewer(OpenOffice/Excel)
• Table viewer

KNIME Workflows Available for Download
http://www.schrodinger.com/knimeworkflows!

Other KNIME Workflows
Cheminformatics
• Cluster by Fingerprint
• Database Analysis
• Maximum Common Substructure Search (MCS)
• Select Diverse Molecules
• Similarity Search
• Substructure Search
Docking and Post-Processing
• Docking and Scoring
• Ensemble Docking
• Loop Over Docking Parameters
• Protein Preparation and Glide Grid Generation
• Validate Docking Parameters
• Virtual Screening
• SiteMap
Pharmacophore Modeling
• Phase Hypothesis Identification
• Phase Screening
• Shape Screening
Molecular Dynamics:
• Desmond Simulation
Molecular Mechanics
• Compare Conformational Search Methods
• Conformational Search and Post-Processing
Quantum mechanics
• Conformational Search and QM Refinement
• ESP Charges
• Jaguar pKa
• Quantum Mechanical Properties
• Semi-empirical Optimization
Library Design
• Library Enumeration
Protein Modeling
• Induced Fit Docking Protocol
• Model Building
Workbench
• Group By Use-cases
• Group Looper
• Unpivot
Real World Examples
• Binding Site Shape Clustering
• Sitemap and Glide Grid Generation
• Vendor Database Preparation
Labs
• Glide Grid Writer
• Parameter Flow Variable Use-cases
• Run Maestro 1:1 Use-cases
General tools
• Chemistry External Tool Use-cases
• Ensure Molecule Title Uniqueness
• Output Column Structure Option Philosophy
• Protein Structure Alignment
• Python Script Node Use-cases
• Run Maestro Command Node Use-cases
• Run PyMOL
• Split and Align Multimers
• Webservice
• Workflows in the Current Workspace
http://www.schrodinger.com/knimeworkflows/
!
Schrödinger nodes ►
• Chemistry tool nodes
• Python nodes
• Row iterator loop start
• Look up and add vs. Joiner node
• Miscellaneous nodes: Compare ligands, Set molecule title

48
Intermediate
KNIME!workbench!
!-!GUI ! !!!!!!!!!!►!
!-!Nodes ! !!!!!!!!!►!
!
Schrödinger!extensions!
!-!Specificities ! !!!!!!!!►!
!-!Schrödinger!nodes ! !!!!!!►!
◄ Get!started!
►!Advanced!functionalities!
KNIME workbench GUI ◄
• Preferences
• Advanced node functionalities
• Errors, warnings and Console information
• Flow variables and workflow variables
• Metanodes
• Memory limit
• Tips and tricks

Preferences > KNIME
• Directory for temporary files (See also Schrödinger preferences ►)
• KNIME GUI- disable the node reset, deletion and reconnection
confirmation
• KNIME GUI- Console view log level: recommended to change to
INFO. Example of information provided by Schrödinger nodes ►

Advanced node functionalities
• Hovering over an input connector tells you what the node takes as input (eg Molecules in
Maestro, SMILES or SD format)
• Hovering over an output connector reports the number of rows and columns in the output table
• Comment a workflow: Node pop-up menu > Node name and description
• Data table > change the renderer

Errors, warnings and Console information
• Popup-menu > View Std output/error
• Warning sign above the node status
when the node completed with potential
errors
• Console information:
INFO HierarchicalClusteringNodeModel Preparing input file '/tmp/HierarchicalClustering_in_423741.mat'
INFO HierarchicalClusteringNodeModel Finished preparing file time 0.35 seconds
INFO HierarchicalClusteringNodeModel 10:42:45 11.17.2009:
Running cmdline[0]='=/usr/local/schro-latest/utilities/canvasHC -im HierarchicalClustering_in_4116794508031023741.mat -ot
HierarchicalClustering_in_4116794508031023741.tree -og HierarchicalClustering_in_4116794508031023741.csv -linkage
schrodinger -n 123'
INFO HierarchicalClusteringNodeModel Completed time 1.626 seconds
INFO HierarchicalClusteringNodeModel Preparing output
INFO HierarchicalClusteringNodeModel Finished preparing output: time 0.06 seconds
INFO LocalNodeExecutionJob Hierarchical Clustering (from Matrix) 0:2:50 End execute (2 secs)

Flow variables and workflow variables
• The Flow variables are used pass data between nodes on top of the connections.
• In the flow variable tab or the configuration panel for a couple of nodes:
• Global variables can also be set: with the Java snippet node ►
Or in the Workflow project repository select the workflow and Workflow variables in the pop-up
menu.
See also Schrödinger specificities ► and nodes to edit variables ►

Metanodes
• To hide the complexity and organize a workflow
• Chose the number and type of input/output
• The metanodes open up in new tabs

Memory limit
• Check the memory limit: Help > About Knime > Installation details > Configuration and search for
a line starting with "eclipse.vmargs=-Xmx" (close to the top).
• Increase the memory allocated to KNIME:
– $SCHRODINGER\knime -maxHeap 4096m
– knime –Xmx4096m (as last option in the command line)
– in $SCHRODINGER\knime-v*\bin\*\knime.ini: change -Xmx1024M into 2048M (or higher on
64 bit)
• The error message usually contain "Java heap space“ when there is a KNIME is running out of
memory.
• Preferences > General > Show heap status and use the garbage collector.
• Knime and Schrödinger tools (eg Canvas) don't compete for memory.

Tips and tricks
• Copy and paste some nodes to a specific place:
Select, copy the nodes (Ctrl+C), right click where you want to paste the
nodes and select Paste in the pop-up menu.
Using Ctrl+V instead the nodes will be pasted a little below the original ones.
• The keyboard shortcuts for items on the menus are listed as usual with the
menu item. In File > Preferences > General > Keys you can view all the key
bindings to commands, modify the bindings, and create your own shortcuts.
• All the branches can be run at the same time using the GUI toolbar Execute
all executable nodes button. See also Cancel all running nodes.

Known issues
• If you can´t save the workflow with a Java heap space error try to disconnect
the last node or run the garbage collector.
KNIME workbench GUI ►
• Report designer
• Global variables
• Batch execution
• Tips and tricks

KNIME workbench node ◄
• KNIME.com Labs nodes
• Scripting and run a third party tool
• Java snippet use-cases
• Manipulate the table row IDs using the RowID node
• Aggregation using the GroupBy node
• Miscellaneous nodes: Interactive table, Math formula, CDK Sketcher
• Plotting facilities
• Looping functionalities- Basics
• Model building nodes

KNIME.com Labs nodes
• Pipeline Pilot Connector (other way around?)
• Web Service client, etc
Specific update site: http://labs.knime.org/

Scripting and run a third party tool
• Java snippet ►
• Jython and Schrödinger Python nodes ►
• Perl scripting
• External tool and Schrödinger Chemistry external tool nodes ►
• Run Maestro commands ►

Java snippet use-cases
• Duplicate numeric or string columns
• Create a new column from scratch (eg a tag)
• Combine columns (and flow variables) but use the Combiner node for
simple tasks
eg return "prefix-"+$$FlowVar$$+"_ref_"+$Col1$;
• Add a row index (see also Set MAE index)
See the corresponding
workflow example.
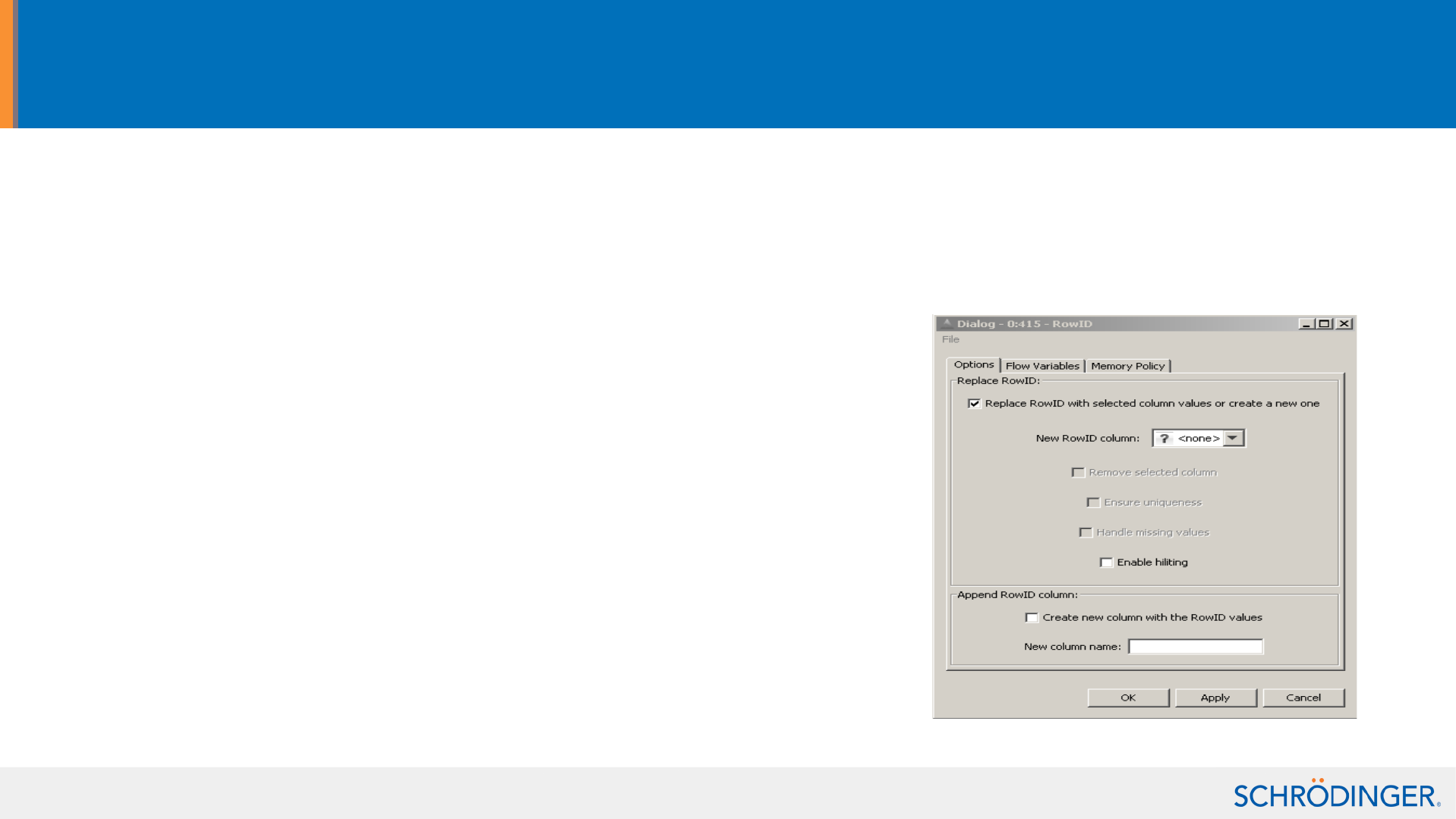
Manipulate the table row IDs using the RowID node
• Use data table column values as row IDs and store Row IDs in a column.
Use-cases:
– before transposing a data table
– Set the labels to be used by the Plotter node
• Ensure row ID uniqueness
– eg for Canvas tools before creating a matrix)

Aggregation using the GroupBy node
Some of the aggregation methods:
- first, last
- max, min
- Mean
- Sum
- Concatenate
- (unique) count
- List
- Set

Miscellaneous nodes
• Interactive table: Find & Find Next
equivalent to the Schrödinger Text viewer node
that have more functionalities
• Math formula
• CDK Structure sketcher
or Marvin sketch
(free of charge from Infocom)

Plotting facilities
• Data Views: Plotter, Histogram…
• Mining > Scoring: Enrichment plotter, ROC curve
• Advanced capabilities available in KNIME Report designer
►

Looping facilities- Basics
• Loop start … Loop end
• Inject and extract variables
• TableRow/Column to and from variables
• Prebuilt protocols
• Schrödinger node: Row iterator loop start ►

Model building nodes
+ Future Canvas nodes
(already some prototypes)

KNIME workbench nodes ►
• Edit variables and advanced looping functionalities
• Hilite functionalities
• Database nodes
• Miscellaneous useful nodes

Schrödinger extensions specificities ◄
• Schrödinger preferences
• Start-up script options
• Access to the flow variables

Schrödinger Preferences
• A specific scratch directory can be specified for Schrödinger nodes
• Delete temporary files after a node successfully executes
Toggled off to run the calculation through the command line again

Start-up script options
To pass user/machine/OS-specific parameters
-maxHeap Maximum heap size eg 2048 for 2G
-maxThreads Maximum working threads
-tempDir Schrödinger extensions temporary directory
-defaultHost Default host
-deleteTempFiles true/false Delete temporary files on or off.
-ooCmd <value> Excel / Open Office Spreadsheet command
eg oocalc
or C:\Program Files (x86)\Microsoft Office\Office12\EXCEL.EXE
And more (see knime –h message)

Access to the flow variables
• Access to the flow variables in the Flow variable tab
including the Chemistry external tools nodes (using %flow_n%)
• Variable based Glide grid reader

Schrödinger nodes ◄
• Chemistry tool nodes
• Python nodes
• Row iterator loop start
• Look up and add vs. Joiner node
• Miscellaneous nodes: Compare ligands, Set molecule title, Get PDB

Chemistry external tool nodes
• Input/output types:
– Maestro, SD, mol2, Smiles
– Double, Integer
– String, Text
– Sequence, alignment
– FingerPrint, Canvas Matrix
– Phase Hypothesis, Glide Grid

Python nodes
• Input/output connectors: 0:1,
1:1, 1:2, 2:2
• Schrödinger’s APIs
• Possibility to include third
party APIs

Row iterator loop start

Look up and add vs. Joiner node
• Take advantage only 1 column, concatenate columns
The Joiner node is the easiest way to concatenate columns when the table have
the same number of rows and same rowIDs.

Miscellaneous nodes
• Get PDB: easy way to get one or several structures
use the | symbol as a delimiter for the list of codes
• Set molecule title
• Compare ligands: the modes are First only, both, either

Schrödinger nodes ►
• Simple workflow examples
• Workflow development support for customers
• Automatic protein preparation
• Scientifically relevant application of the workflow examples
• Interactive work with Knime using the HiLite functionalities
• Use a workflow again

Advanced functionalities
KNIME!workbench!
!-!GUI ! !!!!!!!!!!►!
!-!Nodes ! !!!!!!!!!►!
!
Schrödinger!extensions!
!-!Advanced!workflow!examples ! !!!►!
◄ Intermediate!

KNIME workbench GUI ◄
• Report designer
• Global variables
• Batch execution
• Tips and tricks

Report designer
• From knime.com but free of charge. Included in our distribution
• Include To report node(s) in the workflow (can’t be in metanodes) and switch to the
Report designer mode

Report designer- template mode

Report designer- Canvas 2D renderer
The structures can be shown in a report using Canvas 2D renderer using the following procedure:
• 1. In the workflow, add a MAE-to-smiles node and a To report node.
• 2. In Reporting mode, in the Layout tab, add a table to the report (drag and drop from the Data
set view).
• 3. Insert in the "[smiles]" cell (Table- detail row) an Image widget from the Report Items list.
• 4. Configure the widget (using "Edit" on the widget), select "Dynamic image", and press "Select
Image Data..." to select the source column (which should be the Smiles column). Delete "[smiles]"
if you want just the image and no SMILES. You may want to alter the size of the cell by dragging
the border vertically and horizontally if necessary.
• 5. Change the size of the image to something like 300x300, which is done by editing the Data set
view (right click -> Edit -> Parameters), and changing (or creating new Parameters typed as
integer if they don't exist yet) the knime-image-height and knime-image-width parameters.
• 6. Check the view in the Preview tab

Global variables
• In the Workflow project list, right-click on the workflow, under Workflow variables
Batch execution
• $SCHRODINGER/knime -batch –reset –nosplash -nosave
-workflowFile=<path>/<wkf>.zip or -workflowDir=<path>/<workspace>/<wkf>
• Alter some settings -option=nodeNumber,valueName,value,type
-option=7,filename,"/tmp/new-molprops.csv",String (int, double or String)
Find the node number in the configuration panel header (add the metanode numbers)
eg 123/456/78 for the node 78 in the metanode 456 in the metanode 123
Find the option name in the workspace directory: <workflow>/node_name(#7)/node.xml eg:
<config key="DataURL">
<entry key="array-size" type="xint" value="1" />
<entry key="0" type="xstring" value="/C:/serotonin_unique.sdf" />
-option=2,DataURL\0,"file:/tmp/new-input.mae",String
When the input is an array
• Pass some variables: -workflow.variable=name,value,type (int, double or String)
• Workflows can be run from Maestro using a simple Python script wrapper

Tips and tricks
• Rearrange the panels
• Workflow Meta-Infos
• Try to open a workflow modified with a newer version of KNIME alter the 2
following lines of the file
<workspace>/<workflow>/workflow.knime:
<entry key="created_by" type="xstring" value="2.0.3.0021120"/>
<entry key="version" type="xstring" value="2.0.0"/>

KNIME workbench nodes ◄
• Edit variables and advanced looping functionalities
• HiLite functionalities
• Database nodes
• Miscellaneous useful nodes

Edit variables and advanced looping functionalities
• Extract variable (data) and Inject variable (data)
• TableRow to Variable (use the first row), Variable to
TableRow and Variable to TableColumn

Hilite!functionalities!
• HiLite filter and HiLite collector nodes
• Color, Size and Shape Manager/Appender nodes

Database nodes
• See simple examples (not on the Workflow page yet)

Time series support
• See simple examples (not on the Workflow page yet)

Miscellaneous nodes: Cell splitter, Numeric binner
• Reference row filter, Reference column filter, Nominal value row filter
• Missing values
• Create collection column and Split collection
• Text manipulation: String replacer, Case converter, Cell splitter
• Row sampling, Partitioning, Shuffle
• Numeric binner

KNIME workflow examples ◄
• KNIME workflow page
• Workflow development support for customers
• Workflow example presentation

Homology.modeling.
• Model!building!and!refinement!!!!!
Library.design.
• Library!enumeration!
!!!!!!!!!!!including!a!Run!Maestro!1:1!prototype!
Real.World.Examples.
• Vendor!database!preparation!
General.tools.
• Python!script!node!use-cases!
!!!!!!!!!!!!including!a!Run!PyMOL!prototype!
• Chemistry!external!tool!node!use-cases!
• Run!maestro!command!node!use-cases!
• Output!column!structure!option!philosophy!
KNIME.workbench.
• Workflows!in!the!current!workspace!!
KNIME.workbench-.looper.
• Group!Looper!
!
Simplest,!most.exciting,!new!and!improved!workflows
KNIME workflow page - http:/www.schrodinger.com/knimeworkflows/
Cheminformatics.
• Substructure!Search!
• Clustering,!diversity!selection,!similarity!search.
• Database!analysis!!!!
• Maximum!Common!Substructure!
Docking.and.post-processing.
• Protein!preparation!and!Glide!grid!generation!
• Docking!and!scoring,!Virtual!screening,!Ensemble!docking,!Induced!Fit!
Docking!
• Loop!over!docking!parameters!
• Validate!docking!parameters!
Pharmacophore.modeling.
• Phase!Shape!screening!!!!
• Phase!hypothesis!identification!
• Phase!database!screening!
Molecular.Mechanics.
• Compare!conformational!search!methods!
Quantum.mechanics.
• Conformational!search!and!QM!optimization!
Using!the!Report!designer!

Workflow development support for customers
Combine or expand the workflow examples
• Hierarchical clustering and diverse compounds from each cluster
Waiting for new nodes to be developed
• CombiGlide library enumeration, MacroModel coordinate scan (now available)
• PCA on per residue interactions (Chemistry external tool node)
• Distance measurement in protein and run script in Maestro, descriptor calculation, create protein mutants
(Python script node)
Specific workflows
• Various MacroModel protocols using the Python node for accessing advanced functionalities (eg contraints)
• Prime MM-GB/SA on a set of complexes (ligand detection, flexible residues)
• Simplifying compound docking with KNIME, Dr. Robert Happel, Boehringer Ingelheim, Vienna
http://www.schrodinger.com/seminarprior/19/26/
• Cris Guimaraes MM-GB/SA paper reproduction and improvements
http://www.schrodinger.com/Download.php?type=seminarentry&type2=slides&ident=105
• Protein preparation protocol

Real World Case Study: Binding Site Clustering and Ensemble Docking

Real life applications...

Workflow example presentation
Feel free to request this other presentation including:
• Simple examples
• More advanced examples from the Workflow page
• Scientifically relevant applications

Schrödinger KNIME Extensions
KNIME.Extensions.Product.Manager: .Jean-Christophe!Mozziconacci!!
!!!!!!!!!!!!([email protected]).
Vice.President.of.Technology: . ..Volker!Eyrich.
Main.Developer: . ..Ravikiran!Kuppuraj!
.......and!the!PyDev!development!team!
QA: . .....Simon!Foucher!
Workflow.examples: ! !Tanvi!Bhola!
Technical.Support: . .Katalin!Phimister,!Pavel!Golubkov!.
Marketing: . ...Jarred!Yacob!
